Connexin 43 Gene Ablation Does Not Alter Human Pluripotent Stem Cell Germ Lineage Specification
- PMID: 35053163
- PMCID: PMC8773696
- DOI: 10.3390/biom12010015
Connexin 43 Gene Ablation Does Not Alter Human Pluripotent Stem Cell Germ Lineage Specification
Abstract
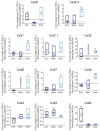
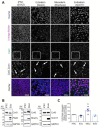
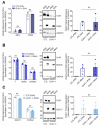
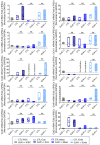
During embryonic germ layer development, cells communicate with each other and their environment to ensure proper lineage specification and tissue development. Connexin (Cx) proteins facilitate direct cell-cell communication through gap junction channels. While previous reports suggest that gap junctional intercellular communication may contribute to germ layer formation, there have been limited comprehensive expression analyses or genetic ablation studies on Cxs during human pluripotent stem cell (PSC) germ lineage specification. We screened the mRNA profile and protein expression patterns of select human Cx isoforms in undifferentiated human induced pluripotent stem cells (iPSCs), and after directed differentiation into the three embryonic germ lineages: ectoderm, definitive endoderm, and mesoderm. Transcript analyses by qPCR revealed upregulation of Cx45 and Cx62 in iPSC-derived ectoderm; Cx45 in mesoderm; and Cx30.3, Cx31, Cx32, Cx36, Cx37, and Cx40 in endoderm relative to control human iPSCs. Generated Cx43 (GJA1) CRISPR-Cas9 knockout iPSCs successfully differentiated into cells of all three germ layers, suggesting that Cx43 is dispensable during directed iPSC lineage specification. Furthermore, qPCR screening of select Cx transcripts in our GJA1-/- iPSCs showed no significant Cx upregulation in response to the loss of Cx43 protein. Future studies will reveal possible compensation by additional Cxs, suggesting targets for future CRISPR-Cas9 ablation studies in human iPSC lineage specification.
Keywords: CRISPR-Cas9 gene ablation; connexin channels; differentiation; germ lineage specification; human pluripotent stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

