Human iPSC-Cardiomyocytes as an Experimental Model to Study Epigenetic Modifiers of Electrophysiology
- PMID: 35053315
- PMCID: PMC8774228
- DOI: 10.3390/cells11020200
Human iPSC-Cardiomyocytes as an Experimental Model to Study Epigenetic Modifiers of Electrophysiology
Abstract
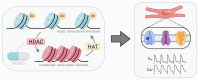
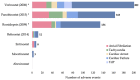
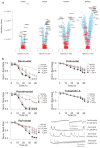
The epigenetic landscape and the responses to pharmacological epigenetic regulators in each human are unique. Classes of epigenetic writers and erasers, such as histone acetyltransferases, HATs, and histone deacetylases, HDACs, control DNA acetylation/deacetylation and chromatin accessibility, thus exerting transcriptional control in a tissue- and person-specific manner. Rapid development of novel pharmacological agents in clinical testing-HDAC inhibitors (HDACi)-targets these master regulators as common means of therapeutic intervention in cancer and immune diseases. The action of these epigenetic modulators is much less explored for cardiac tissue, yet all new drugs need to be tested for cardiotoxicity. To advance our understanding of chromatin regulation in the heart, and specifically how modulation of DNA acetylation state may affect functional electrophysiological responses, human-induced pluripotent stem-cell-derived cardiomyocyte (hiPSC-CM) technology can be leveraged as a scalable, high-throughput platform with ability to provide patient-specific insights. This review covers relevant background on the known roles of HATs and HDACs in the heart, the current state of HDACi development, applications, and any adverse cardiac events; it also summarizes relevant differential gene expression data for the adult human heart vs. hiPSC-CMs along with initial transcriptional and functional results from using this new experimental platform to yield insights on epigenetic control of the heart. We focus on the multitude of methodologies and workflows needed to quantify responses to HDACis in hiPSC-CMs. This overview can help highlight the power and the limitations of hiPSC-CMs as a scalable experimental model in capturing epigenetic responses relevant to the human heart.
Keywords: DNA acetylation; HDAC inhibitors; cardiac electrophysiology; cardiac epigenetics; histone acetyltransferases; histone deacetylases; human iPSC-CMs.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Rakyan V.K., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., et al. DNA Methylation Profiling of the Human Major Histocompatibility Complex: A Pilot Study for the Human Epigenome Project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

