Revisiting the Non-Coding Nature of Pospiviroids
- PMID: 35053381
- PMCID: PMC8773695
- DOI: 10.3390/cells11020265
Revisiting the Non-Coding Nature of Pospiviroids
Abstract
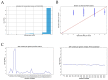
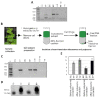
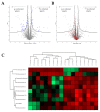
Viroids are small, circular, highly structured pathogens that infect a broad range of plants, causing economic losses. Since their discovery in the 1970s, they have been considered as non-coding pathogens. In the last few years, the discovery of other RNA entities, similar in terms of size and structure, that were shown to be translated (e.g., cirRNAs, precursors of miRNA, RNA satellites) as well as studies showing that some viroids are located in ribosomes, have reignited the idea that viroids may be translated. In this study, we used advanced bioinformatic analysis, in vitro experiments and LC-MS/MS to search for small viroid peptides of the PSTVd. Our results suggest that in our experimental conditions, even though the circular form of PSTVd is found in ribosomes, no produced peptides were identified. This indicates that the presence of PSTVd in ribosomes is most probably not related to peptide production but rather to another unknown function that requires further study.
Keywords: PSTVd; cirRNA; mass spectrometry; non-coding RNA; translation; viroid.
Conflict of interest statement
The authors declare that there is no conflict of interest. This work does not contain any human/animal experiments, and no personal information was used.
Figures




References
-
- Parsyan A. Translation and Its Regulation in Cancer Biology and Medicine. Springer Publishing; Cham, Switzerland: 2014.
Publication types
MeSH terms
Substances
Grants and funding
- 2018ΣΕ01300000/Emblematic Action for Research in the Cretan Agrofood sector: Four Institutions, Four References'-AGRO4Crete
- 2018ΣE01300000/Development of national research network to 'Grape' of the national scale of the public investments program of the GSRT.
- MIS 5002802/The Greek Research Infrastructure for Personalized Medicine (pMED-GR)" which is implemented under the Action "Reinforcement of the Research and Innovation Infrastructure", funded by the Operational Program "Competitiveness, Entrepreneurship and Innovatio
- 155219-17/Natural Sciences and Engineering Research Council of Canada
- 0/Grant from the Université de Sherbrooke

