Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics
- PMID: 35054423
- PMCID: PMC8780579
- DOI: 10.3390/life12010030
Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics
Abstract
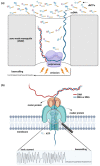
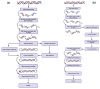
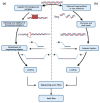
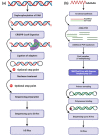
Although next-generation sequencing (NGS) technology revolutionized sequencing, offering a tremendous sequencing capacity with groundbreaking depth and accuracy, it continues to demonstrate serious limitations. In the early 2010s, the introduction of a novel set of sequencing methodologies, presented by two platforms, Pacific Biosciences (PacBio) and Oxford Nanopore Sequencing (ONT), gave birth to third-generation sequencing (TGS). The innovative long-read technologies turn genome sequencing into an ease-of-handle procedure by greatly reducing the average time of library construction workflows and simplifying the process of de novo genome assembly due to the generation of long reads. Long sequencing reads produced by both TGS methodologies have already facilitated the decipherment of transcriptional profiling since they enable the identification of full-length transcripts without the need for assembly or the use of sophisticated bioinformatics tools. Long-read technologies have also provided new insights into the field of epitranscriptomics, by allowing the direct detection of RNA modifications on native RNA molecules. This review highlights the advantageous features of the newly introduced TGS technologies, discusses their limitations and provides an in-depth comparison regarding their scientific background and available protocols as well as their potential utility in research and clinical applications.
Keywords: PacBio sequencing; direct RNA sequencing; epigenomics; epitranscriptomics; long-read sequencing; metagenomics; nanopore sequencing; single-molecule real-time sequencing; targeted DNA sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

