Mining the Wheat Grain Proteome
- PMID: 35054899
- PMCID: PMC8775872
- DOI: 10.3390/ijms23020713
Mining the Wheat Grain Proteome
Abstract
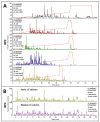
Bread wheat is the most widely cultivated crop worldwide, used in the production of food products and a feed source for animals. Selection tools that can be applied early in the breeding cycle are needed to accelerate genetic gain for increased wheat production while maintaining or improving grain quality if demand from human population growth is to be fulfilled. Proteomics screening assays of wheat flour can assist breeders to select the best performing breeding lines and discard the worst lines. In this study, we optimised a robust LC-MS shotgun quantitative proteomics method to screen thousands of wheat genotypes. Using 6 cultivars and 4 replicates, we tested 3 resuspension ratios (50, 25, and 17 µL/mg), 2 extraction buffers (with urea or guanidine-hydrochloride), 3 sets of proteases (chymotrypsin, Glu-C, and trypsin/Lys-C), and multiple LC settings. Protein identifications by LC-MS/MS were used to select the best parameters. A total 8738 wheat proteins were identified. The best method was validated on an independent set of 96 cultivars and peptides quantities were normalised using sample weights, an internal standard, and quality controls. Data mining tools found particularly useful to explore the flour proteome are presented (UniProt Retrieve/ID mapping tool, KEGG, AgriGO, REVIGO, and Pathway Tools).
Keywords: LC–MS/MS; Triticum aestivum; data mining; normalisation; protease; shotgun proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- International Wheat Genome Sequencing Consortium (IWGSC) IWGSC RefSeq Principal Investigators. Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N., iIWGSC Whole-Genome Assembly Principal Investigators. Pozniak C.J., et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:eaar7191. doi: 10.1126/science.aar7191. - DOI - PubMed
-
- Bonomi F., Iametti S., Mamone G., Ferranti P. The Performing Protein: Beyond Wheat Proteomics? Cereal Chem. 2013;90:358–366. doi: 10.1094/CCHEM-11-12-0161-FI. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

