Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use
- PMID: 35054945
- PMCID: PMC8776057
- DOI: 10.3390/ijms23020764
Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use
Abstract
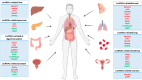
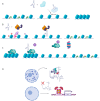
Deep whole genome and transcriptome sequencing have highlighted the importance of an emerging class of non-coding RNA longer than 200 nucleotides (i.e., long non-coding RNAs (lncRNAs)) that are involved in multiple cellular processes such as cell differentiation, embryonic development, and tissue homeostasis. Cancer is a prime example derived from a loss of homeostasis, primarily caused by genetic alterations both in the genomic and epigenetic landscape, which results in deregulation of the gene networks. Deregulation of the expression of many lncRNAs in samples, tissues or patients has been pointed out as a molecular regulator in carcinogenesis, with them acting as oncogenes or tumor suppressor genes. Herein, we summarize the distinct molecular regulatory mechanisms described in literature in which lncRNAs modulate carcinogenesis, emphasizing epigenetic and genetic alterations in particular. Furthermore, we also reviewed the current strategies used to block lncRNA oncogenic functions and their usefulness as potential therapeutic targets in several carcinomas.
Keywords: cancer disease; lncRNAs; therapeutic drugs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Gonzalez D., et al. The GENCODE v7 Catalogue of Human Long Non-Coding RNAs: Analysis of Their Structure, Evolution and Expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

