Computational Design of Miniproteins as SARS-CoV-2 Therapeutic Inhibitors
- PMID: 35055023
- PMCID: PMC8776159
- DOI: 10.3390/ijms23020838
Computational Design of Miniproteins as SARS-CoV-2 Therapeutic Inhibitors
Abstract
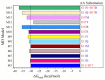
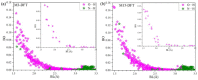
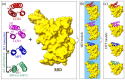
A rational therapeutic strategy is urgently needed for combating SARS-CoV-2 infection. Viral infection initiates when the SARS-CoV-2 receptor-binding domain (RBD) binds to the ACE2 receptor, and thus, inhibiting RBD is a promising therapeutic for blocking viral entry. In this study, the structure of lead antiviral candidate binder (LCB1), which has three alpha-helices (H1, H2, and H3), is used as a template to design and simulate several miniprotein RBD inhibitors. LCB1 undergoes two modifications: structural modification by truncation of the H3 to reduce its size, followed by single and double amino acid substitutions to enhance its binding with RBD. We use molecular dynamics (MD) simulations supported by ab initio density functional theory (DFT) calculations. Complete binding profiles of all miniproteins with RBD have been determined. The MD investigations reveal that the H3 truncation results in a small inhibitor with a -1.5 kcal/mol tighter binding to RBD than original LCB1, while the best miniprotein with higher binding affinity involves D17R or E11V + D17R mutation. DFT calculations provide atomic-scale details on the role of hydrogen bonding and partial charge distribution in stabilizing the minibinder:RBD complex. This study provides insights into general principles for designing potential therapeutics for SARS-CoV-2.
Keywords: SARS-CoV-2; density functional calculation; miniprotein inhibitors modification; molecular dynamics simulation; therapeutics design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Tian J.-H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Logue J., Portnoff A.D., Norton J., Guebre-Xabier M., et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021;12:372. doi: 10.1038/s41467-020-20653-8. - DOI - PMC - PubMed
-
- Kang Y.-F., Sun C., Zhuang Z., Yuan R.-Y., Zheng Q., Li J.-P., Zhou P.-P., Chen X.-C., Liu Z., Zhang X., et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano. 2021;15:2738–2752. doi: 10.1021/acsnano.0c08379. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

