Common and Distinctive Intercellular Communication Patterns in Human Obstructive and Nonobstructive Hypertrophic Cardiomyopathy
- PMID: 35055131
- PMCID: PMC8780670
- DOI: 10.3390/ijms23020946
Common and Distinctive Intercellular Communication Patterns in Human Obstructive and Nonobstructive Hypertrophic Cardiomyopathy
Abstract
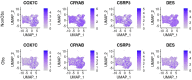
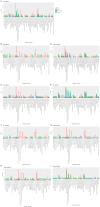
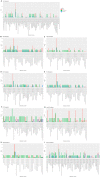
Hypertrophic Cardiomyopathy (HCM) is a common inherited disorder characterized by unexplained left ventricular hypertrophy with or without left ventricular outflow tract (LVOT) obstruction. Single-nuclei RNA-sequencing (snRNA-seq) of both obstructive and nonobstructive HCM patient samples has revealed alterations in communication between various cell types, but no direct and integrated comparison between the two HCM phenotypes has been reported. We performed a bioinformatic analysis of HCM snRNA-seq datasets from obstructive and nonobstructive patient samples to identify differentially expressed genes and distinctive patterns of intercellular communication. Differential gene expression analysis revealed 37 differentially expressed genes, predominantly in cardiomyocytes but also in other cell types, relevant to aging, muscle contraction, cell motility, and the extracellular matrix. Intercellular communication was generally reduced in HCM, affecting the extracellular matrix, growth factor binding, integrin binding, PDGF binding, and SMAD binding, but with increases in adenylate cyclase binding, calcium channel inhibitor activity, and serine-threonine kinase activity in nonobstructive HCM. Increases in neuron to leukocyte and dendritic cell communication, in fibroblast to leukocyte and dendritic cell communication, and in endothelial cell communication to other cell types, largely through changes in the expression of integrin-β1 and its cognate ligands, were also noted. These findings indicate both common and distinct physiological mechanisms affecting the pathogenesis of obstructive and nonobstructive HCM and provide opportunities for the personalized management of different HCM phenotypes.
Keywords: Hypertrophic Cardiomyopathy; dendritic cells; integrin-β1; left ventricular outflow tract obstruction; single-nucleus RNA-sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
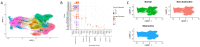




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

