Paving Therapeutic Avenues for FOXG1 Syndrome: Untangling Genotypes and Phenotypes from a Molecular Perspective
- PMID: 35055139
- PMCID: PMC8780739
- DOI: 10.3390/ijms23020954
Paving Therapeutic Avenues for FOXG1 Syndrome: Untangling Genotypes and Phenotypes from a Molecular Perspective
Abstract
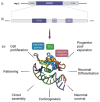
Development of the central nervous system (CNS) depends on accurate spatiotemporal control of signaling pathways and transcriptional programs. Forkhead Box G1 (FOXG1) is one of the master regulators that play fundamental roles in forebrain development; from the timing of neurogenesis, to the patterning of the cerebral cortex. Mutations in the FOXG1 gene cause a rare neurodevelopmental disorder called FOXG1 syndrome, also known as congenital form of Rett syndrome. Patients presenting with FOXG1 syndrome manifest a spectrum of phenotypes, ranging from severe cognitive dysfunction and microcephaly to social withdrawal and communication deficits, with varying severities. To develop and improve therapeutic interventions, there has been considerable progress towards unravelling the multi-faceted functions of FOXG1 in the neurodevelopment and pathogenesis of FOXG1 syndrome. Moreover, recent advances in genome editing and stem cell technologies, as well as the increased yield of information from high throughput omics, have opened promising and important new avenues in FOXG1 research. In this review, we provide a summary of the clinical features and emerging molecular mechanisms underlying FOXG1 syndrome, and explore disease-modelling approaches in animals and human-based systems, to highlight the prospects of research and possible clinical interventions.
Keywords: FOXG1; FOXG1 syndrome; Rett syndrome; brain development; disease modelling; hiPSCs; neurodevelopmental disorders; organoids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
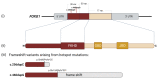
- Kortum F., Das S., Flindt M., Morris-Rosendahl D.J., Stefanova I., Goldstein A., Horn D., Klopocki E., Kluger G., Martin P., et al. The Core FOXG1 Syndrome Phenotype Consists of Postnatal Microcephaly, Severe Mental Retardation, Absent Language, Dyskinesia, and Corpus Callosum Hypogenesis. J. Med. Genet. 2011;48:396–406. doi: 10.1136/jmg.2010.087528. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

