Phenotypic Selection of Dairy Cattle Infected with Bovine Leukemia Virus Demonstrates Immunogenetic Resilience through NGS-Based Genotyping of BoLA MHC Class II Genes
- PMID: 35056052
- PMCID: PMC8779071
- DOI: 10.3390/pathogens11010104
Phenotypic Selection of Dairy Cattle Infected with Bovine Leukemia Virus Demonstrates Immunogenetic Resilience through NGS-Based Genotyping of BoLA MHC Class II Genes
Abstract
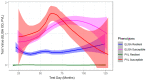
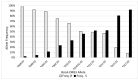
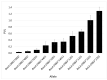
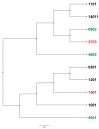
Characterization of the bovine leukocyte antigen (BoLA) DRB3 gene has shown that specific alleles associate with susceptibility or resilience to the progression of bovine leukemia virus (BLV), measured by proviral load (PVL). Through surveillance of multi-farm BLV eradication field trials, we observed differential phenotypes within seropositive cows that persist from months to years. We sought to develop a multiplex next-generation sequencing workflow (NGS-SBT) capable of genotyping 384 samples per run to assess the relationship between BLV phenotype and two BoLA genes. We utilized longitudinal results from milk ELISA screening and subsequent blood collections on seropositive cows for PVL determination using a novel BLV proviral load multiplex qPCR assay to phenotype the cows. Repeated diagnostic observations defined two distinct phenotypes in our study population, ELISA-positive cows that do not harbor detectable levels of provirus and those who do have persistent proviral loads. In total, 565 cows from nine Midwest dairy farms were selected for NGS-SBT, with 558 cows: 168 BLV susceptible (ELISA-positive/PVL-positive) and 390 BLV resilient (ELISA-positive/PVL-negative) successfully genotyped. Three BoLA-DRB3 alleles, including one novel allele, were shown to associate with disease resilience, *009:02, *044:01, and *048:02 were found at rates of 97.5%, 86.5%, and 90.3%, respectively, within the phenotypically resilient population. Alternatively, DRB3*015:01 and *027:03, both known to associate with disease progression, were found at rates of 81.1% and 92.3%, respectively, within the susceptible population. This study helps solidify the immunogenetic relationship between BoLA-DRB3 alleles and BLV infection status of these two phenotypic groupings of US dairy cattle.
Keywords: BoLA; ELISA; MHC Class II; bovine leukemia virus; dairy science; disease resilience; immunogenetics; molecular diagnostics; proviral load; qPCR; sequence-based typing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ferrer J.F., Marshak R.R., Abt D.A., Kenyon S.J. Persistent lymphocytosis in cattle: Its cause, nature and relation to lymphosarcoma. Ann. Rech. Vét. 1978;9:851–857. - PubMed
-
- Johnson R., Gibson C.D., Kaneene J.B. Bovine leukemia virus: A herd-based control strategy. Prev. Vet. Med. 1985;3:339–349. doi: 10.1016/0167-5877(85)90011-X. - DOI
-
- Furtado M.D.S.B.S., Andrade R.G., Romanelli L.C.F., Ribeiro M.A., Ribas J.G., Torres E.B., Barbosa-Stancioli E.F., Proietti A.B.D.F.C., Martins M.L. Monitoring the HTLV-1 proviral load in the peripheral blood of asymptomatic carriers and patients with HTLV-associated myelopathy/tropical spastic paraparesis from a Brazilian cohort: ROC curve analysis to establish the threshold for risk disease. J. Med Virol. 2012;84:664–671. doi: 10.1002/jmv.23227. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

