Repositioning of Etravirine as a Potential CK1ε Inhibitor by Virtual Screening
- PMID: 35056065
- PMCID: PMC8778358
- DOI: 10.3390/ph15010008
Repositioning of Etravirine as a Potential CK1ε Inhibitor by Virtual Screening
Abstract
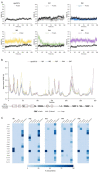
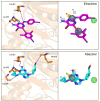
CK1ε is a key regulator of WNT/β-catenin and other pathways that are linked to tumor progression; thus, CK1ε is considered a target for the development of antineoplastic therapies. In this study, we performed a virtual screening to search for potential CK1ε inhibitors. First, we characterized the dynamic noncovalent interactions profiles for a set of reported CK1ε inhibitors to generate a pharmacophore model, which was used to identify new potential inhibitors among FDA-approved drugs. We found that etravirine and abacavir, two drugs that are approved for HIV infections, can be repurposed as CK1ε inhibitors. The interaction of these drugs with CK1ε was further examined by molecular docking and molecular dynamics. Etravirine and abacavir formed stable complexes with the target, emulating the binding behavior of known inhibitors. However, only etravirine showed high theoretical binding affinity to CK1ε. Our findings provide a new pharmacophore for targeting CK1ε and implicate etravirine as a CK1ε inhibitor and antineoplastic agent.
Keywords: CK1ε; abacavir; cancer; drug repurposing; etravirine; pharmacophore model.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

