Occasional paternal inheritance of the germline-restricted chromosome in songbirds
- PMID: 35058355
- PMCID: PMC8794876
- DOI: 10.1073/pnas.2103960119
Occasional paternal inheritance of the germline-restricted chromosome in songbirds
Abstract
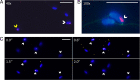
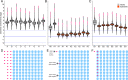
Songbirds have one special accessory chromosome, the so-called germline-restricted chromosome (GRC), which is only present in germline cells and absent from all somatic tissues. Earlier work on the zebra finch (Taeniopygia guttata castanotis) showed that the GRC is inherited only through the female line-like the mitochondria-and is eliminated from the sperm during spermatogenesis. Here, we show that the GRC has the potential to be paternally inherited. Confocal microscopy using GRC-specific fluorescent in situ hybridization probes indicated that a considerable fraction of sperm heads (1 to 19%) in zebra finch ejaculates still contained the GRC. In line with these cytogenetic data, sequencing of ejaculates revealed that individual males from two families differed strongly and consistently in the number of GRCs in their ejaculates. Examining a captive-bred male hybrid of the two zebra finch subspecies (T. g. guttata and T. g. castanotis) revealed that the mitochondria originated from a castanotis mother, whereas the GRC came from a guttata father. Moreover, analyzing GRC haplotypes across nine castanotis matrilines, estimated to have diverged for up to 250,000 y, showed surprisingly little variability among GRCs. This suggests that a single GRC haplotype has spread relatively recently across all examined matrilines. A few diagnostic GRC mutations that arose since this inferred spreading suggest that the GRC has continued to jump across matriline boundaries. Our findings raise the possibility that certain GRC haplotypes could selfishly spread through the population via occasional paternal transmission, thereby outcompeting other GRC haplotypes that were limited to strict maternal inheritance, even if this was partly detrimental to organismal fitness.
Keywords: elimination efficiency; germline-restricted chromosome; paternal spillover; selfish DNA; zebra finch.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Pigozzi M. I., Solari A. J., Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosome Res. 6, 105–113 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

