Recurrent mutations in topoisomerase IIα cause a previously undescribed mutator phenotype in human cancers
- PMID: 35058360
- PMCID: PMC8795545
- DOI: 10.1073/pnas.2114024119
Recurrent mutations in topoisomerase IIα cause a previously undescribed mutator phenotype in human cancers
Abstract
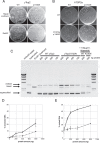
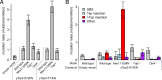
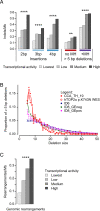
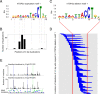
Topoisomerases nick and reseal DNA to relieve torsional stress associated with transcription and replication and to resolve structures such as knots and catenanes. Stabilization of the yeast Top2 cleavage intermediates is mutagenic in yeast, but whether this extends to higher eukaryotes is less clear. Chemotherapeutic topoisomerase poisons also elevate cleavage, resulting in mutagenesis. Here, we describe p.K743N mutations in human topoisomerase hTOP2α and link them to a previously undescribed mutator phenotype in cancer. Overexpression of the orthologous mutant protein in yeast generated a characteristic pattern of 2- to 4-base pair (bp) duplications resembling those in tumors with p.K743N. Using mutant strains and biochemical analysis, we determined the genetic requirements of this mutagenic process and showed that it results from trapping of the mutant yeast yTop2 cleavage complex. In addition to 2- to 4-bp duplications, hTOP2α p.K743N is also associated with deletions that are absent in yeast. We call the combined pattern of duplications and deletions ID_TOP2α. All seven tumors carrying the hTOP2α p.K743N mutation showed ID_TOP2α, while it was absent from all other tumors examined (n = 12,269). Each tumor with the ID_TOP2α signature had indels in several known cancer genes, which included frameshift mutations in tumor suppressors PTEN and TP53 and an activating insertion in BRAF. Sequence motifs found at ID_TOP2α mutations were present at 80% of indels in cancer-driver genes, suggesting that ID_TOP2α mutagenesis may contribute to tumorigenesis. The results reported here shed further light on the role of topoisomerase II in genome instability.
Keywords: cancer; duplications; indel mutational signature; topoisomerase II; yeast.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: S.G.R., A.B., and one of the reviewers of this manuscript, Professor E.R., participated in the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium, which comprised over 700 researchers from around the world. S.G.R., A.B., and Professor E.R. did not directly work or publish together.
Figures

References
-
- Wang J. C., Moving one DNA double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 (1998). - PubMed
-
- Corbett K. D., Berger J. M., Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

