Proteomic cellular signatures of kinase inhibitor-induced cardiotoxicity
- PMID: 35058449
- PMCID: PMC8776854
- DOI: 10.1038/s41597-021-01114-3
Proteomic cellular signatures of kinase inhibitor-induced cardiotoxicity
Abstract
Drug Toxicity Signature Generation Center (DToxS) at the Icahn School of Medicine at Mount Sinai is one of the centers for the NIH Library of Integrated Network-Based Cellular Signatures (LINCS) program. Its key aim is to generate proteomic and transcriptomic signatures that can predict cardiotoxic adverse effects of kinase inhibitors approved by the Food and Drug Administration. Towards this goal, high throughput shotgun proteomics experiments (308 cell line/drug combinations +64 control lysates) have been conducted. Using computational network analyses, these proteomic data can be integrated with transcriptomic signatures, generated in tandem, to identify cellular signatures of cardiotoxicity that may predict kinase inhibitor-induced toxicity and enable possible mitigation. Both raw and processed proteomics data have passed several quality control steps and been made publicly available on the PRIDE database. This broad protein kinase inhibitor-stimulated human cardiomyocyte proteomic data and signature set is valuable for prediction of drug toxicities.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
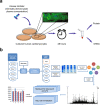
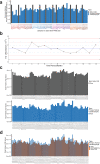


Dataset use reported in
- doi: 10.1038/s41467-020-18396-7
- doi: 10.1038/s41597-021-01008-4
References
-
- Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1S10OD025047/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54HG008098/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01DK118222/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54 HL127624/HL/NHLBI NIH HHS/United States
- R01GM072853/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54HL127624/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30NS046593/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 GM112415/GM/NIGMS NIH HHS/United States
- P30 NS046593/NS/NINDS NIH HHS/United States
- R01 DK118222/DK/NIDDK NIH HHS/United States
- R01GM112415/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54 HG008098/HG/NHGRI NIH HHS/United States
- S10 OD025047/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

