Cooperative allostery and structural dynamics of streptavidin at cryogenic- and ambient-temperature
- PMID: 35058563
- PMCID: PMC8776744
- DOI: 10.1038/s42003-021-02903-7
Cooperative allostery and structural dynamics of streptavidin at cryogenic- and ambient-temperature
Abstract
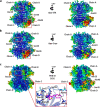
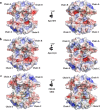
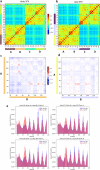
Multimeric protein assemblies are abundant in nature. Streptavidin is an attractive protein that provides a paradigm system to investigate the intra- and intermolecular interactions of multimeric protein complexes. Also, it offers a versatile tool for biotechnological applications. Here, we present two apo-streptavidin structures, the first one is an ambient temperature Serial Femtosecond X-ray crystal (Apo-SFX) structure at 1.7 Å resolution and the second one is a cryogenic crystal structure (Apo-Cryo) at 1.1 Å resolution. These structures are mostly in agreement with previous structural data. Combined with computational analysis, these structures provide invaluable information about structural dynamics of apo streptavidin. Collectively, these data further reveal a novel cooperative allostery of streptavidin which binds to substrate via water molecules that provide a polar interaction network and mimics the substrate biotin which displays one of the strongest affinities found in nature.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
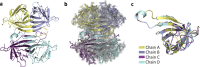




References
-
- Lim, K. H., Huang, H., Pralle, A. & Park, S. Engineered streptavidin monomer and dimer with improved stability and function. Biochemistry10.1021/bi2010366 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

