Formation, Development, and Cross-Species Interactions in Biofilms
- PMID: 35058893
- PMCID: PMC8764401
- DOI: 10.3389/fmicb.2021.757327
Formation, Development, and Cross-Species Interactions in Biofilms
Abstract
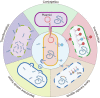
Biofilms, which are essential vectors of bacterial survival, protect microbes from antibiotics and host immune attack and are one of the leading causes that maintain drug-resistant chronic infections. In nature, compared with monomicrobial biofilms, polymicrobial biofilms composed of multispecies bacteria predominate, which means that it is significant to explore the interactions between microorganisms from different kingdoms, species, and strains. Cross-microbial interactions exist during biofilm development, either synergistically or antagonistically. Although research into cross-species biofilms remains at an early stage, in this review, the important mechanisms that are involved in biofilm formation are delineated. Then, recent studies that investigated cross-species cooperation or synergy, competition or antagonism in biofilms, and various components that mediate those interactions will be elaborated. To determine approaches that minimize the harmful effects of biofilms, it is important to understand the interactions between microbial species. The knowledge gained from these investigations has the potential to guide studies into microbial sociality in natural settings and to help in the design of new medicines and therapies to treat bacterial infections.
Keywords: biofilm; cross-species interactions; horizontal gene transfer; metabolic interactions; quorum sensing.
Copyright © 2022 Luo, Wang, Sun, Liu and Xin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

