INHBA is a Prognostic Biomarker and Correlated With Immune Cell Infiltration in Cervical Cancer
- PMID: 35058963
- PMCID: PMC8764128
- DOI: 10.3389/fgene.2021.705512
INHBA is a Prognostic Biomarker and Correlated With Immune Cell Infiltration in Cervical Cancer
Abstract
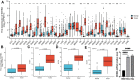
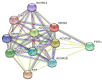
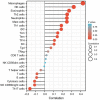
Background: Inhibin A (INHBA), a member of the TGF-β superfamily, has been shown to be differentially expressed in various cancer types and is associated with prognosis. However, its role in cervical cancer remains unclear. Methods: We aimed to demonstrate the relationship between INHBA expression and pan-cancer using The Cancer Genome Atlas (TCGA) database. Next, we validated INHBA expression in cervical cancer using the Gene Expression Omnibus (GEO) database, including GSE7803, GSE63514, and GSE9750 datasets. Enrichment analysis of INHBA was performed using the R package "clusterProfiler." We analyzed the association between immune infiltration level and INHBA expression in cervical cancer using the single-sample gene set enrichment analysis (ssGSEA) method by the R package GSVA. We explored the association between INHBA expression and prognosis using the R package "survival". Results: Pan-cancer data analysis showed that INHBA expression was elevated in 19 tumor types, including cervical cancer. We further confirmed that INHBA expression was higher in cervical cancer samples from GEO database and cervical cancer cell lines than in normal cervical cells. Survival prognosis analysis indicated that higher INHBA expression was significantly associated with reduced Overall Survival (p = 0.001), disease Specific Survival (p = 0.006), and Progression Free Interval (p = 0.001) in cervical cancer and poorer prognosis in other tumors. GSEA and infiltration analysis showed that INHBA expression was significantly associated with tumor progression and some types of immune infiltrating cells. Conclusion: INHBA was highly expressed in cervical cancer and was significantly associated with poor prognosis. Meanwhile, it was correlated with immune cell infiltration and could be used as a promising prognostic target for cervical cancer.
Keywords: INHBA; biomarker; cervical cancer; immune infiltration; prognosis.
Copyright © 2022 Zhao, Yi, Ma and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Comprehensive analysis of inhibin-β A as a potential biomarker for gastrointestinal tract cancers through bioinformatics approaches.Sci Rep. 2025 Jan 7;15(1):1090. doi: 10.1038/s41598-024-72679-3. Sci Rep. 2025. PMID: 39774945 Free PMC article.
-
Comprehensive Analysis to Identify SPP1 as a Prognostic Biomarker in Cervical Cancer.Front Genet. 2022 Jan 4;12:732822. doi: 10.3389/fgene.2021.732822. eCollection 2021. Front Genet. 2022. PMID: 35058964 Free PMC article.
-
Increased Expression of INHBA Is Correlated With Poor Prognosis and High Immune Infiltrating Level in Breast Cancer.Front Bioinform. 2022 Mar 24;2:729902. doi: 10.3389/fbinf.2022.729902. eCollection 2022. Front Bioinform. 2022. PMID: 36304286 Free PMC article.
-
A pan-cancer analysis targeting the oncogenic role of ATP-binding cassette transporter A1 in human tumors.Front Oncol. 2025 Apr 14;15:1513992. doi: 10.3389/fonc.2025.1513992. eCollection 2025. Front Oncol. 2025. PMID: 40297807 Free PMC article. Review.
-
Thirteen years of clusterProfiler.Innovation (Camb). 2024 Oct 21;5(6):100722. doi: 10.1016/j.xinn.2024.100722. eCollection 2024 Nov 4. Innovation (Camb). 2024. PMID: 39529960 Free PMC article. Review. No abstract available.
Cited by
-
COL10A1 Facilitates Prostate Cancer Progression by Interacting With INHBA to Activate the PI3K/AKT Pathway.J Cell Mol Med. 2024 Dec;28(23):e70249. doi: 10.1111/jcmm.70249. J Cell Mol Med. 2024. PMID: 39656597 Free PMC article.
-
Comprehensive analysis of inhibin-β A as a potential biomarker for gastrointestinal tract cancers through bioinformatics approaches.Sci Rep. 2025 Jan 7;15(1):1090. doi: 10.1038/s41598-024-72679-3. Sci Rep. 2025. PMID: 39774945 Free PMC article.
-
Defining Melanoma Immune Biomarkers-Desert, Excluded, and Inflamed Subtypes-Using a Gene Expression Classifier Reflecting Intratumoral Immune Response and Stromal Patterns.Biomolecules. 2024 Jan 31;14(2):171. doi: 10.3390/biom14020171. Biomolecules. 2024. PMID: 38397409 Free PMC article.
-
Cellograph: a semi-supervised approach to analyzing multi-condition single-cell RNA-sequencing data using graph neural networks.BMC Bioinformatics. 2024 Jan 15;25(1):25. doi: 10.1186/s12859-024-05641-9. BMC Bioinformatics. 2024. PMID: 38221640 Free PMC article.
-
Macrophage membrane (MMs) camouflaged near-infrared (NIR) responsive bone defect area targeting nanocarrier delivery system (BTNDS) for rapid repair: promoting osteogenesis via phototherapy and modulating immunity.J Nanobiotechnology. 2024 Mar 1;22(1):87. doi: 10.1186/s12951-024-02351-5. J Nanobiotechnology. 2024. PMID: 38429776 Free PMC article.
References
-
- Babion I., Snoek B., Novianti P., Jaspers A., van Trommel N. V., Heideman D., et al. (2018). Triage of High-Risk Hpv-Positive Women in Population-Based Screening by Mirna Expression Analysis in Cervical Scrapes; a Feasibility Study. Clin. Epigenetics 10, 1. 10.1186/s13148-018-0509-9 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

