findMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM
- PMID: 35059213
- PMCID: PMC8733886
- DOI: 10.1107/S2052252521011088
findMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM
Abstract
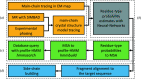
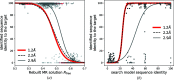
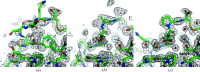
Although experimental protein-structure determination usually targets known proteins, chains of unknown sequence are often encountered. They can be purified from natural sources, appear as an unexpected fragment of a well characterized protein or appear as a contaminant. Regardless of the source of the problem, the unknown protein always requires characterization. Here, an automated pipeline is presented for the identification of protein sequences from cryo-EM reconstructions and crystallographic data. The method's application to characterize the crystal structure of an unknown protein purified from a snake venom is presented. It is also shown that the approach can be successfully applied to the identification of protein sequences and validation of sequence assignments in cryo-EM protein structures.
Keywords: SIMBAD; bioinformatics; cryo-EM; findMySequence; neural networks; protein sequences; protein structures; structure determination.
© Grzegorz Chojnowski et al. 2022.
Figures


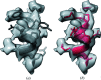

References
-
- Amazonas, D. R., Portes-Junior, J. A., Nishiyama-Jr, M. Y., Nicolau, C. A., Chalkidis, H. M., Mourão, R. H. V., Grazziotin, F. G., Rokyta, D. R., Gibbs, H. L., Valente, R. H., Junqueira-de-Azevedo, I. L. M. & Moura-da-Silva, A. M. (2018). J. Proteomics, 181, 60–72. - PubMed
LinkOut - more resources
Full Text Sources

