Genomic Surveillance Enables the Identification of Co-infections With Multiple SARS-CoV-2 Lineages in Equatorial Guinea
- PMID: 35059385
- PMCID: PMC8763705
- DOI: 10.3389/fpubh.2021.818401
Genomic Surveillance Enables the Identification of Co-infections With Multiple SARS-CoV-2 Lineages in Equatorial Guinea
Abstract
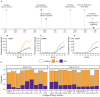
COVID-19 disease caused by SARS-CoV-2 represents an ongoing global public health emergency. Rapid identification of emergence, evolution, and spread of SARS-CoV-2 variants of concern (VOC) would enable timely and tailored responses by public health decision-making bodies. Yet, global disparities in current SARS-CoV-2 genomic surveillance activities reveal serious geographical gaps. Here, we discuss the experiences and lessons learned from the SARS-CoV-2 monitoring and surveillance program at the Public Health Laboratory on Bioko Island, Equatorial Guinea that was implemented as part of the national COVID-19 response and monitoring activities. We report how three distinct SARS-CoV-2 variants have dominated the epidemiological situation in Equatorial Guinea since March 2020. In addition, a case of co-infection of two SARS-CoV-2 VOC, Beta and Delta, in a clinically asymptomatic and fully COVID-19 vaccinated man living in Equatorial Guinea is presented. To our knowledge, this is the first report of a person co-infected with Beta and Delta VOC globally. Rapid identification of co-infections is relevant since these might provide an opportunity for genetic recombination resulting in emergence of novel SARS-CoV-2 lineages with enhanced transmission or immune evasion potential.
Keywords: Central-Africa; SARS-CoV-2; co-infection; genomic surveillance; variant of concern.
Copyright © 2022 Hosch, Mpina, Nyakurungu, Borico, Obama, Ovona, Wagner, Rubin, Vickos, Milang, Ayekaba, Phiri, Daubenberger and Schindler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Scaling, Up Genomic Sequencing in Africa,. WHO. Regional Office for Africa. Available online at: https://www.afro.who.int/news/scaling-genomic-sequencing-africa (accessed November 13, 2021).
-
- Vega-Magaña N, Sánchez-Sánchez R, Hernández-Bello J, Venancio-Landeros AA, Peña-Rodríguez M, Vega-Zepeda RA, et al. . RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: a screening strategy to identify variants with clinical impact. Front Cell Infect Microbiol. (2021) 11:434. 10.3389/FCIMB.2021.672562/BIBTEX - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

