A Xanthomonas transcription activator-like effector is trapped in nonhost plants for immunity
- PMID: 35059629
- PMCID: PMC8760140
- DOI: 10.1016/j.xplc.2021.100249
A Xanthomonas transcription activator-like effector is trapped in nonhost plants for immunity
Abstract
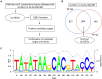
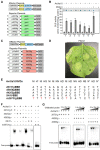
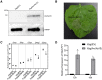
Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial leaf blight in rice, delivers transcription activator-like effector (TALE) proteins into host cells to activate susceptibility or resistance (R) genes that promote disease or immunity, respectively. Nonhost plants serve as potential reservoirs of R genes; consequently, nonhost R genes may trap TALEs to trigger an immune response. In this study, we screened 17 Xoo TALEs for their ability to induce a hypersensitive response (HR) in the nonhost plant Nicotiana benthamiana (Nb); only AvrXa10 elicited an HR when transiently expressed in Nb. The HR generated by AvrXa10 required both the central repeat region and the activation domain, suggesting a specific interaction between AvrXa10 and a potential R-like gene in nonhost plants. Evans blue staining and ion leakage measurements confirmed that the AvrXa10-triggered HR was a form of cell death, and the transient expression of AvrXa10 in Nb induced immune responses. Genes targeted by AvrXa10 in the Nb genome were identified by transcriptome profiling and prediction of effector binding sites. Using several approaches (in vivo reporter assays, electrophoretic mobility-shift assays, targeted designer TALEs, and on-spot gene silencing), we confirmed that AvrXa10 targets NbZnFP1, a C2H2-type zinc finger protein that resides in the nucleus. Functional analysis indicated that overexpression of NbZnFP1 and its rice orthologs triggered cell death in rice protoplasts. An NbZnFP1 ortholog was also identified in tomato and was specifically activated by AvrXa10. These results demonstrate that NbZnFP1 is a nonhost R gene that traps AvrXa10 to promote plant immunity in Nb.
Keywords: AvrXa10; Xanthomonas oryzae pv. oryzae; hypersensitive response; nonhost plant; zinc finger protein.
© 2021 The Authors.
Figures




Similar articles
-
TALEs as double-edged swords in plant-pathogen interactions: Progress, challenges, and perspectives.Plant Commun. 2022 May 9;3(3):100318. doi: 10.1016/j.xplc.2022.100318. Epub 2022 Mar 25. Plant Commun. 2022. PMID: 35576155 Free PMC article. Review.
-
AvrXa7-Xa7 mediated defense in rice can be suppressed by transcriptional activator-like effectors TAL6 and TAL11a from Xanthomonas oryzae pv. oryzicola.Mol Plant Microbe Interact. 2014 Sep;27(9):983-95. doi: 10.1094/MPMI-09-13-0279-R. Mol Plant Microbe Interact. 2014. Retraction in: Mol Plant Microbe Interact. 2014 Dec;27(12):1413. PMID: 25105804 Retracted.
-
A transcription activator-like effector from Xanthomonas oryzae pv. oryzicola elicits dose-dependent resistance in rice.Mol Plant Pathol. 2017 Jan;18(1):55-66. doi: 10.1111/mpp.12377. Epub 2016 Apr 21. Mol Plant Pathol. 2017. PMID: 26821568 Free PMC article.
-
A Transcription Activator-Like Effector Tal7 of Xanthomonas oryzae pv. oryzicola Activates Rice Gene Os09g29100 to Suppress Rice Immunity.Sci Rep. 2017 Jul 11;7(1):5089. doi: 10.1038/s41598-017-04800-8. Sci Rep. 2017. PMID: 28698641 Free PMC article.
-
TALE-induced cell death executors: an origin outside immunity?Trends Plant Sci. 2022 Jun;27(6):536-548. doi: 10.1016/j.tplants.2021.11.003. Epub 2021 Dec 16. Trends Plant Sci. 2022. PMID: 34924289 Free PMC article. Review.
Cited by
-
Tal6b/AvrXa27A, a hidden TALE targeting the susceptibility gene OsSWEET11a and the resistance gene Xa27 in rice.Plant Commun. 2024 Feb 12;5(2):100721. doi: 10.1016/j.xplc.2023.100721. Epub 2023 Sep 20. Plant Commun. 2024. PMID: 37735868 Free PMC article.
-
Genome-Wide Identification and Analysis of GATA Gene Family in Dendrobium officinale Under Methyl Jasmonate and Salt Stress.Plants (Basel). 2025 May 22;14(11):1576. doi: 10.3390/plants14111576. Plants (Basel). 2025. PMID: 40508251 Free PMC article.
-
TALEs as double-edged swords in plant-pathogen interactions: Progress, challenges, and perspectives.Plant Commun. 2022 May 9;3(3):100318. doi: 10.1016/j.xplc.2022.100318. Epub 2022 Mar 25. Plant Commun. 2022. PMID: 35576155 Free PMC article. Review.
-
BrWRKY8: a key regulatory factor involved in delaying postharvest leaf senescence of Pakchoi (Brassica rapa subsp. chinensis) by 2,4-epibrassinolide.Hortic Res. 2025 Jan 6;12(4):uhaf004. doi: 10.1093/hr/uhaf004. eCollection 2025 Apr. Hortic Res. 2025. PMID: 40078720 Free PMC article.
-
Two TAL Effectors of Xanthomonas citri pv. malvacearum Induce Water Soaking by Activating GhSWEET14 Genes in Cotton.Mol Plant Pathol. 2025 Jan;26(1):e70053. doi: 10.1111/mpp.70053. Mol Plant Pathol. 2025. PMID: 39825471 Free PMC article.
References
-
- Asai S., Yoshioka H. Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 2009;22:619–629. - PubMed
-
- Boch J., Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

