Advances in proteome-wide analysis of plant lysine acetylation
- PMID: 35059632
- PMCID: PMC8760137
- DOI: 10.1016/j.xplc.2021.100266
Advances in proteome-wide analysis of plant lysine acetylation
Abstract
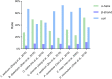
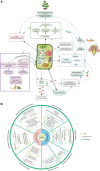
Lysine acetylation (LysAc) is a conserved and important post-translational modification (PTM) that plays a key role in plant physiological and metabolic processes. Based on advances in Lys-acetylated protein immunoenrichment and mass-spectrometric technology, LysAc proteomics studies have been performed in many species. Such studies have made substantial contributions to our understanding of plant LysAc, revealing that Lys-acetylated histones and nonhistones are involved in a broad spectrum of plant cellular processes. Here, we present an extensive overview of recent research on plant Lys-acetylproteomes. We provide in-depth insights into the characteristics of plant LysAc modifications and the mechanisms by which LysAc participates in cellular processes and regulates metabolism and physiology during plant growth and development. First, we summarize the characteristics of LysAc, including the properties of Lys-acetylated sites, the motifs that flank Lys-acetylated lysines, and the dynamic alterations in LysAc among different tissues and developmental stages. We also outline a map of Lys-acetylated proteins in the Calvin-Benson cycle and central carbon metabolism-related pathways. We then introduce some examples of the regulation of plant growth, development, and biotic and abiotic stress responses by LysAc. We discuss the interaction between LysAc and Nα-terminal acetylation and the crosstalk between LysAc and other PTMs, including phosphorylation and succinylation. Finally, we propose recommendations for future studies in the field. We conclude that LysAc of proteins plays an important role in the regulation of the plant life cycle.
Keywords: PTM crosstalk; lysine acetylproteomes; modified characteristics; plant growth and development; stress responses.
© 2021 The Author(s).
Figures



References
-
- Armbruster L., Linster E., Boyer J.B., Brunje A., Eirich J., Stephan I., Bienvenut W.V., Weidenhausen J., Meinnel T., Hell R., et al. NAA50 is an enzymatically active Nα-acetyltransferase that is crucial for development and regulation of stress responses. Plant Physiol. 2020;183:1502–1516. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

