Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants
- PMID: 35059633
- PMCID: PMC8760139
- DOI: 10.1016/j.xplc.2021.100267
Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants
Abstract
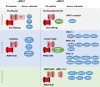
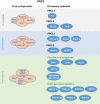
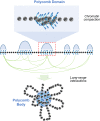
The evolutionary conserved Polycomb Group (PcG) repressive system comprises two central protein complexes, PcG repressive complex 1 (PRC1) and PRC2. These complexes, through the incorporation of histone modifications on chromatin, have an essential role in the normal development of eukaryotes. In recent years, a significant effort has been made to characterize these complexes in the different kingdoms, and despite there being remarkable functional and mechanistic conservation, some key molecular principles have diverged. In this review, we discuss current views on the function of plant PcG complexes. We compare the composition of PcG complexes between animals and plants, highlight the role of recently identified plant PcG accessory proteins, and discuss newly revealed roles of known PcG partners. We also examine the mechanisms by which the repression is achieved and how these complexes are recruited to target genes. Finally, we consider the possible role of some plant PcG proteins in mediating local and long-range chromatin interactions and, thus, shaping chromatin 3D architecture.
Keywords: PRC1; PRC2; Polycomb Group; chromatin organization; gene repression; histone modifications.
© 2021 The Author(s).
Figures


References
-
- Ariel F., Lucero L., Christ A., Mammarella M.F., Jegu T., Veluchamy A., Mariappan K., Latrasse D., Blein T., Liu C., et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell. 2020;77:1055–1065.e4. - PubMed
-
- Bastola D.R., Pethe V.V., Winicov I. Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 1998;38:1123–1135. - PubMed

