Protocol for single-cell isolation and genome amplification of environmental microbial eukaryotes for genomic analysis
- PMID: 35059648
- PMCID: PMC8760549
- DOI: 10.1016/j.xpro.2021.100968
Protocol for single-cell isolation and genome amplification of environmental microbial eukaryotes for genomic analysis
Abstract
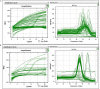
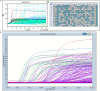
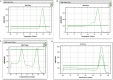
We describe environmental microbial eukaryotes (EMEs) sample collection, single-cell isolation, lysis, and genome amplification, followed by the rDNA amplification and OTU screening for recovery of high-quality species-specific genomes for de novo assembly. These protocols are part of our pipeline that also includes bioinformatic methods. The pipeline and its application on a wide range of phyla of different sample complexity are described in our complementary paper. In addition, this protocol describes optimized lysis, genome amplification, and OTU screening steps of the pipeline. For complete details on the use and execution of this protocol, please refer to Ciobanu et al. (2021).
Keywords: Cell isolation; Environmental sciences; Flow Cytometry/Mass Cytometry; Genomics; Microbiology; Molecular Biology; Sequence analysis; Sequencing; Single Cell.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- BD Influx™ Cell Sorter User’s Guide. (2011) bdbiosciences.com 23-11543-00 Rev. 01 4/2011.
-
- Lazarus K.L., Benny G.L., Ho H.M., Smith M.E. Phylogenetic systematics of Syncephalis (Zoopagales, Zoopagomycotina), a genus of ubiquitous mycoparasites. Mycologia. 2017;109:333–349. - PubMed

