In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy
- PMID: 35061543
- PMCID: PMC8782450
- DOI: 10.1126/sciadv.abj6901
In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy
Abstract
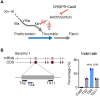
Hemophilia is a hereditary disease that remains incurable. Although innovative treatments such as gene therapy or bispecific antibody therapy have been introduced, substantial unmet needs still exist with respect to achieving long-lasting therapeutic effects and treatment options for inhibitor patients. Antithrombin (AT), an endogenous negative regulator of thrombin generation, is a potent genome editing target for sustainable treatment of patients with hemophilia A and B. In this study, we developed and optimized lipid nanoparticles (LNPs) to deliver Cas9 mRNA along with single guide RNA that targeted AT in the mouse liver. The LNP-mediated CRISPR-Cas9 delivery resulted in the inhibition of AT that led to improvement in thrombin generation. Bleeding-associated phenotypes were recovered in both hemophilia A and B mice. No active off-targets, liver-induced toxicity, and substantial anti-Cas9 immune responses were detected, indicating that the LNP-mediated CRISPR-Cas9 delivery was a safe and efficient approach for hemophilia therapy.
Figures





References
-
- Peters R., Harris T., Advances and innovations in haemophilia treatment. Nat. Rev. Drug Discov. 17, 493–508 (2018). - PubMed
-
- Valentino L. A., Cooper D. L., Goldstein B., Surgical experience with rFVIIa (NovoSeven) in congenital haemophilia A and B patients with inhibitors to factors VIII or IX. Haemophilia 17, 579–589 (2011). - PubMed
-
- van den Berg H. M., Fischer K., Carcao M., Chambost H., Kenet G., Kurnik K., Konigs C., Male C., Santagostino E., Ljung R.; PedNet Study Group , Timing of inhibitor development in more than 1000 previously untreated patients with severe hemophilia A. Blood 134, 317–320 (2019). - PubMed
-
- Oldenburg J., Levy G. G., Emicizumab prophylaxis in hemophilia a with inhibitors. N. Engl. J. Med. 377, 2194–2195 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

