Transient Cell Cycle Induction in Cardiomyocytes to Treat Subacute Ischemic Heart Failure
- PMID: 35061545
- PMCID: PMC9038650
- DOI: 10.1161/CIRCULATIONAHA.121.057641
Transient Cell Cycle Induction in Cardiomyocytes to Treat Subacute Ischemic Heart Failure
Abstract
Background: The regenerative capacity of the heart after myocardial infarction is limited. Our previous study showed that ectopic introduction of 4 cell cycle factors (4F; CDK1 [cyclin-dependent kinase 1], CDK4 [cyclin-dependent kinase 4], CCNB [cyclin B1], and CCND [cyclin D1]) promotes cardiomyocyte proliferation in 15% to 20% of infected cardiomyocytes in vitro and in vivo and improves cardiac function after myocardial infarction in mice.
Methods: Using temporal single-cell RNA sequencing, we aimed to identify the necessary reprogramming stages during the forced cardiomyocyte proliferation with 4F on a single cell basis. Using rat and pig models of ischemic heart failure, we aimed to start the first preclinical testing to introduce 4F gene therapy as a candidate for the treatment of ischemia-induced heart failure.
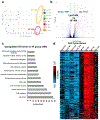
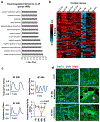
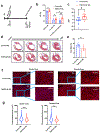
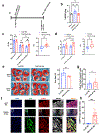
Results: Temporal bulk and single-cell RNA sequencing and further biochemical validations of mature human induced pluripotent stem cell-derived cardiomyocytes treated with either LacZ or 4F adenoviruses revealed full cell cycle reprogramming in 15% of the cardiomyocyte population at 48 hours after infection with 4F, which was associated mainly with sarcomere disassembly and metabolic reprogramming (n=3/time point/group). Transient overexpression of 4F, specifically in cardiomyocytes, was achieved using a polycistronic nonintegrating lentivirus (NIL) encoding 4F; each is driven by a TNNT2 (cardiac troponin T isoform 2) promoter (TNNT2-4Fpolycistronic-NIL). TNNT2-4Fpolycistronic-NIL or control virus was injected intramyocardially 1 week after myocardial infarction in rats (n=10/group) or pigs (n=6-7/group). Four weeks after injection, TNNT2-4Fpolycistronic-NIL-treated animals showed significant improvement in left ventricular ejection fraction and scar size compared with the control virus-treated animals. At 4 months after treatment, rats that received TNNT2-4Fpolycistronic-NIL still showed a sustained improvement in cardiac function and no obvious development of cardiac arrhythmias or systemic tumorigenesis (n=10/group).
Conclusions: This study provides mechanistic insights into the process of forced cardiomyocyte proliferation and advances the clinical feasibility of this approach by minimizing the oncogenic potential of the cell cycle factors owing to the use of a novel transient and cardiomyocyte-specific viral construct.
Keywords: cardiomyopathies; cell cycle; genetic therapy; heart failure; metabolism; sarcomeres.
Conflict of interest statement
Conflict of Interest Disclosures
R.R.E.A.: None. A.M.S.: None. Q.O.: None. X-L.T.: None. M.S.: None. K.M.K.: None. Y.G.: None. Y.H.: None. Y.N.: None. L.M.: None. P.K.L.: None. B.G.H.: None. K.C.: None. R.T.: None. B.M.A.: None. J.S.: None. H.R.J.: None. A.S.: None. Z.I.: None. S.H.: None. D.J.C.: None. A.S.E.: None. K.N.I: Holds equities at Tenaya Therapeutics. D.S.: Holds equities at Tenaya Therapeutics. R.B.: None. T.M.A.M.: Holds equities at Tenaya Therapeutics.
Figures


Comment in
-
At the Brink of Human Therapy to Generate New Myocytes in the Adult Injured Heart.Circulation. 2022 Apr 26;145(17):1356-1358. doi: 10.1161/CIRCULATIONAHA.122.059106. Epub 2022 Apr 25. Circulation. 2022. PMID: 35467953 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

