Control of backbone chemistry and chirality boost oligonucleotide splice switching activity
- PMID: 35061895
- PMCID: PMC9178015
- DOI: 10.1093/nar/gkac018
Control of backbone chemistry and chirality boost oligonucleotide splice switching activity
Erratum in
-
Correction to 'Control of backbone chemistry and chirality boost oligonucleotide splice switching activity'.Nucleic Acids Res. 2023 Apr 24;51(7):3498. doi: 10.1093/nar/gkad185. Nucleic Acids Res. 2023. PMID: 36912093 Free PMC article. No abstract available.
Abstract
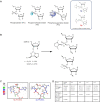
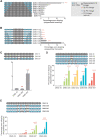
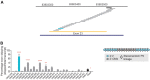
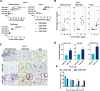
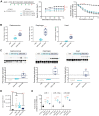
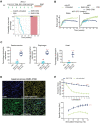
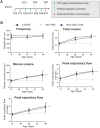
Although recent regulatory approval of splice-switching oligonucleotides (SSOs) for the treatment of neuromuscular disease such as Duchenne muscular dystrophy has been an advance for the splice-switching field, current SSO chemistries have shown limited clinical benefit due to poor pharmacology. To overcome limitations of existing technologies, we engineered chimeric stereopure oligonucleotides with phosphorothioate (PS) and phosphoryl guanidine-containing (PN) backbones. We demonstrate that these chimeric stereopure oligonucleotides have markedly improved pharmacology and efficacy compared with PS-modified oligonucleotides, preventing premature death and improving median survival from 49 days to at least 280 days in a dystrophic mouse model with an aggressive phenotype. These data demonstrate that chemical optimization alone can profoundly impact oligonucleotide pharmacology and highlight the potential for continued innovation around the oligonucleotide backbone. More specifically, we conclude that chimeric stereopure oligonucleotides are a promising splice-switching modality with potential for the treatment of neuromuscular and other genetic diseases impacting difficult to reach tissues such as the skeletal muscle and heart.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Verhaart I.E.C., Aartsma-Rus A.. Therapeutic developments for duchenne muscular dystrophy. Nat. Rev. Neurol. 2019; 15:373–386. - PubMed
-
- Birnkrant D.J., Bushby K., Bann C.M., Apkon S.D., Blackwell A., Brumbaugh D., Case L.E., Clemens P.R., Hadjiyannakis S., Pandya S.et al. .. Diagnosis and management of duchenne muscular dystrophy, part 1: diagnosis, and neuromuscular, rehabilitation, endocrine, and gastrointestinal and nutritional management. Lancet. Neurology. 2018; 17:251–267. - PMC - PubMed
-
- Blake D.J., Weir A., Newey S.E., Davies K.E.. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev. 2002; 82:291–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

