Phosphate-induced resistance to pathogen infection in Arabidopsis
- PMID: 35061924
- PMCID: PMC9303409
- DOI: 10.1111/tpj.15680
Phosphate-induced resistance to pathogen infection in Arabidopsis
Abstract
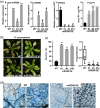
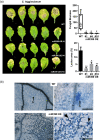
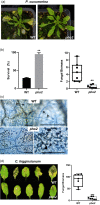
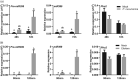
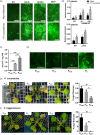
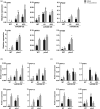
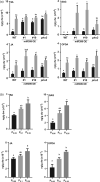
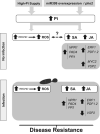
In nature, plants are concurrently exposed to a number of abiotic and biotic stresses. Our understanding of convergence points between responses to combined biotic/abiotic stress pathways remains, however, rudimentary. Here we show that MIR399 overexpression, loss-of-function of PHOSPHATE2 (PHO2), or treatment with high phosphate (Pi) levels is accompanied by an increase in Pi content and accumulation of reactive oxygen species (ROS) in Arabidopsis thaliana. High Pi plants (e.g., miR399 overexpressors, pho2 mutants, and plants grown under high Pi supply) exhibited resistance to infection by necrotrophic and hemibiotrophic fungal pathogens. In the absence of pathogen infection, the expression levels of genes in the salicylic acid (SA)- and jasmonic acid (JA)-dependent signaling pathways were higher in high Pi plants compared to wild-type plants grown under control conditions, which is consistent with increased levels of SA and JA in non-infected high Pi plants. During infection, an opposite regulation in the two branches of the JA pathway (ERF1/PDF1.2 and MYC2/VSP2) occurs in high Pi plants. Thus, while pathogen infection induces PDF1.2 expression in miR399 OE and pho2 plants, VSP2 expression is downregulated by pathogen infection in these plants. This study supports the notion that Pi accumulation promotes resistance to infection by fungal pathogens in Arabidopsis, while providing a basis to better understand interactions between Pi signaling and hormonal signaling pathways for modulation of plant immune responses.
Keywords: Arabidopsis thaliana; Colletotrichum higginsianum; PHOSPHATE 2; Plectosphaerella cucumerina; immune response; jasmonic acid (JA); microRNA399 (miR399); phosphate (Pi); reactive oxygen species (ROS); salicylic acid (SA).
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they do not have competing interests.
Figures
References
-
- Atkinson, N.J. & Urwin, P.E. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. Journal of Experimental Botany, 63, 3523–3544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

