SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters
- PMID: 35062015
- PMCID: PMC8942849
- DOI: 10.1038/s41586-022-04441-6
SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters
Abstract
The recent emergence of B.1.1.529, the Omicron variant1,2, has raised concerns of escape from protection by vaccines and therapeutic antibodies. A key test for potential countermeasures against B.1.1.529 is their activity in preclinical rodent models of respiratory tract disease. Here, using the collaborative network of the SARS-CoV-2 Assessment of Viral Evolution (SAVE) programme of the National Institute of Allergy and Infectious Diseases (NIAID), we evaluated the ability of several B.1.1.529 isolates to cause infection and disease in immunocompetent and human ACE2 (hACE2)-expressing mice and hamsters. Despite modelling data indicating that B.1.1.529 spike can bind more avidly to mouse ACE2 (refs. 3,4), we observed less infection by B.1.1.529 in 129, C57BL/6, BALB/c and K18-hACE2 transgenic mice than by previous SARS-CoV-2 variants, with limited weight loss and lower viral burden in the upper and lower respiratory tracts. In wild-type and hACE2 transgenic hamsters, lung infection, clinical disease and pathology with B.1.1.529 were also milder than with historical isolates or other SARS-CoV-2 variants of concern. Overall, experiments from the SAVE/NIAID network with several B.1.1.529 isolates demonstrate attenuated lung disease in rodents, which parallels preliminary human clinical data.
© 2022. The Author(s).
Conflict of interest statement
M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. The Boon laboratory has received unrelated funding support in sponsored research agreements from AI Therapeutics, AbbVie Inc., GreenLight Biosciences Inc., and Nano targeting & Therapy Biopharma Inc. M.S.S. serves on Advisory boards for Moderna and Ocugen. Y.K. has received unrelated funding support from Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Inc., Shionogi & Co. Ltd, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Shinya Corporation and Fuji Rebio. The A.G.-S. laboratory has received unrelated research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma and Merck. A.G.-S. has paid or equity-based consulting agreements for the following companies: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus, CureLab Oncology, CureLab Veterinary and Pfizer, outside the reported work. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside the reported work. Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays that list Viviana Simon as co-inventor. S.P. has received unrelated research support from BioAge Laboratories and Autonomous Therapeutics Inc. The remaining authors declare no competing interests.
Figures
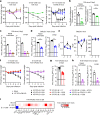
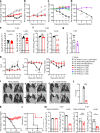
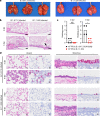
Update of
-
The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters.Res Sq [Preprint]. 2021 Dec 29:rs.3.rs-1211792. doi: 10.21203/rs.3.rs-1211792/v1. Res Sq. 2021. Update in: Nature. 2022 Mar;603(7902):687-692. doi: 10.1038/s41586-022-04441-6. PMID: 34981044 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32 AI007647/AI/NIAID NIH HHS/United States
- P51OD011132/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- 75N93021C00017/AI/NIAID NIH HHS/United States
- U01 AI151810/AI/NIAID NIH HHS/United States
- R01 DK130425/DK/NIDDK NIH HHS/United States
- R01 AI129269/AI/NIAID NIH HHS/United States
- P01 AI060699/AI/NIAID NIH HHS/United States
- HHSN272201400004C/AI/NIAID NIH HHS/United States
- S10 OD030463/OD/NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
- R56 AI147623/AI/NIAID NIH HHS/United States
- 75N93019C00051/AI/NIAID NIH HHS/United States
- R56AI147623/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P51 OD011132/OD/NIH HHS/United States
- HHSN272201700041I/AI/NIAID NIH HHS/United States
- R01 AI157155/AI/NIAID NIH HHS/United States
- R01DK130425/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- 75N93021C00016/AI/NIAID NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- 75N93020F00001/A38/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI129269/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

