Emerging SARS-CoV-2 Genotypes Show Different Replication Patterns in Human Pulmonary and Intestinal Epithelial Cells
- PMID: 35062227
- PMCID: PMC8777977
- DOI: 10.3390/v14010023
Emerging SARS-CoV-2 Genotypes Show Different Replication Patterns in Human Pulmonary and Intestinal Epithelial Cells
Abstract
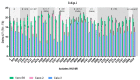
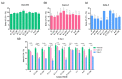
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) quickly spread worldwide following its emergence in Wuhan, China, and hit pandemic levels. Its tremendous incidence favoured the emergence of viral variants. The current genome diversity of SARS-CoV-2 has a clear impact on epidemiology and clinical practice, especially regarding transmission rates and the effectiveness of vaccines. In this study, we evaluated the replication of different SARS-CoV-2 isolates representing different virus genotypes which have been isolated throughout the pandemic. We used three distinct cell lines, including Vero E6 cells originating from monkeys; Caco-2 cells, an intestinal epithelium cell line originating from humans; and Calu-3 cells, a pulmonary epithelium cell line also originating from humans. We used RT-qPCR to replicate different SARS-CoV-2 genotypes by quantifying the virus released in the culture supernatant of infected cells. We found that the different viral isolates replicate similarly in Caco-2 cells, but show very different replicative capacities in Calu-3 cells. This was especially highlighted for the lineages B.1.1.7, B.1.351 and P.1, which are considered to be variants of concern. These results underscore the importance of the evaluation and characterisation of each SARS-CoV-2 isolate in order to establish the replication patterns before performing tests, and of the consideration of the ideal SARS-CoV-2 genotype-cell type pair for each assay.
Keywords: COVID-19; Caco-2 cells; Calu-3 cells; SARS-CoV-2; Vero E6 cells; genotype; isolate; variants; viral culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
SARS-CoV-2 Isolates Show Impaired Replication in Human Immune Cells but Differential Ability to Replicate and Induce Innate Immunity in Lung Epithelial Cells.Microbiol Spectr. 2021 Sep 3;9(1):e0077421. doi: 10.1128/Spectrum.00774-21. Epub 2021 Aug 11. Microbiol Spectr. 2021. PMID: 34378952 Free PMC article.
-
Replication Kinetics of B.1.351 and B.1.1.7 SARS-CoV-2 Variants of Concern Including Assessment of a B.1.1.7 Mutant Carrying a Defective ORF7a Gene.Viruses. 2021 Jun 7;13(6):1087. doi: 10.3390/v13061087. Viruses. 2021. PMID: 34200386 Free PMC article.
-
SARS-CoV-2 variants reveal features critical for replication in primary human cells.PLoS Biol. 2021 Mar 24;19(3):e3001006. doi: 10.1371/journal.pbio.3001006. eCollection 2021 Mar. PLoS Biol. 2021. PMID: 33760807 Free PMC article.
-
SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review.
-
Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models.Virol J. 2022 Apr 26;19(1):76. doi: 10.1186/s12985-022-01802-5. Virol J. 2022. PMID: 35473640 Free PMC article.
Cited by
-
Combinative workflow for mRNA vaccine development.Biochem Biophys Rep. 2024 Jun 28;39:101766. doi: 10.1016/j.bbrep.2024.101766. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39040540 Free PMC article.
-
COVID-19 Clinical Features and Outcomes in Elderly Patients during Six Pandemic Waves.J Clin Med. 2022 Nov 17;11(22):6803. doi: 10.3390/jcm11226803. J Clin Med. 2022. PMID: 36431282 Free PMC article.
-
SARS-CoV-2 in the environment: Contamination routes, detection methods, persistence and removal in wastewater treatment plants.Sci Total Environ. 2023 Jul 10;881:163453. doi: 10.1016/j.scitotenv.2023.163453. Epub 2023 Apr 12. Sci Total Environ. 2023. PMID: 37059142 Free PMC article. Review.
-
Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants.Pharmaceuticals (Basel). 2022 Apr 2;15(4):445. doi: 10.3390/ph15040445. Pharmaceuticals (Basel). 2022. PMID: 35455442 Free PMC article.
-
The SARS-CoV-2 S1 Spike Protein Promotes MAPK and NF-kB Activation in Human Lung Cells and Inflammatory Cytokine Production in Human Lung and Intestinal Epithelial Cells.Microorganisms. 2022 Oct 10;10(10):1996. doi: 10.3390/microorganisms10101996. Microorganisms. 2022. PMID: 36296272 Free PMC article.
References
-
- Chu H., Chan J.F.-W., Yuen T.T.-T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.-Y., Tsang J.O.-L., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. - DOI - PMC - PubMed
-
- Murgolo N., Therien A.G., Howell B., Klein D., Koeplinger K., Lieberman L.A., Adam G.C., Flynn J., McKenna P., Swaminathan G., et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathog. 2021;17:e1009225. doi: 10.1371/journal.ppat.1009225. - DOI - PMC - PubMed
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

