The Thiazole-5-Carboxamide GPS491 Inhibits HIV-1, Adenovirus, and Coronavirus Replication by Altering RNA Processing/Accumulation
- PMID: 35062264
- PMCID: PMC8779516
- DOI: 10.3390/v14010060
The Thiazole-5-Carboxamide GPS491 Inhibits HIV-1, Adenovirus, and Coronavirus Replication by Altering RNA Processing/Accumulation
Abstract
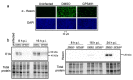
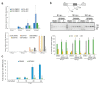
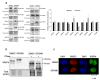
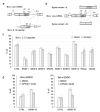
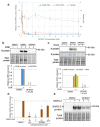
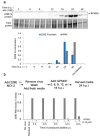
Medicinal chemistry optimization of a previously described stilbene inhibitor of HIV-1, 5350150 (2-(2-(5-nitro-2-thienyl)vinyl)quinoline), led to the identification of the thiazole-5-carboxamide derivative (GPS491), which retained potent anti-HIV-1 activity with reduced toxicity. In this report, we demonstrate that the block of HIV-1 replication by GPS491 is accompanied by a drastic inhibition of viral gene expression (IC50 ~ 0.25 µM), and alterations in the production of unspliced, singly spliced, and multiply spliced HIV-1 RNAs. GPS491 also inhibited the replication of adenovirus and multiple coronaviruses. Low µM doses of GPS491 reduced adenovirus infectious yield ~1000 fold, altered virus early gene expression/viral E1A RNA processing, blocked viral DNA amplification, and inhibited late (hexon) gene expression. Loss of replication of multiple coronaviruses (229E, OC43, SARS-CoV2) upon GPS491 addition was associated with the inhibition of viral structural protein expression and the formation of virus particles. Consistent with the observed changes in viral RNA processing, GPS491 treatment induced selective alterations in the accumulation/phosphorylation/function of splicing regulatory SR proteins. Our study establishes that a compound that impacts the activity of cellular factors involved in RNA processing can prevent the replication of several viruses with minimal effect on cell viability.
Keywords: HIV-1; RNA processing; adenovirus; coronavirus; inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

