Detection and Molecular Characterization of Canine Alphacoronavirus in Free-Roaming Dogs, Bangladesh
- PMID: 35062271
- PMCID: PMC8778797
- DOI: 10.3390/v14010067
Detection and Molecular Characterization of Canine Alphacoronavirus in Free-Roaming Dogs, Bangladesh
Abstract
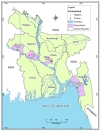
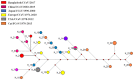
Canine coronavirus (CCoV) is widespread among the dog population and causes gastrointestinal disorders, and even fatal cases. As the zoonotic transmission of viruses from animals to humans has become a worldwide concern nowadays, it is necessary to screen free-roaming dogs for their common pathogens due to their frequent interaction with humans. We conducted a cross-sectional study to detect and characterize the known and novel Corona, Filo, Flavi, and Paramyxoviruses in free-roaming dogs in Bangladesh. Between 2009-10 and 2016-17, we collected swab samples from 69 dogs from four districts of Bangladesh, tested using RT-PCR and sequenced. None of the samples were positive for Filo, Flavi, and Paramyxoviruses. Only three samples (4.3%; 95% CI: 0.9-12.2) tested positive for Canine Coronavirus (CCoV). The CCoV strains identified were branched with strains of genotype CCoV-II with distinct distances. They are closely related to CCoVs from the UK, China, and other CoVs isolated from different species, which suggests genetic recombination and interspecies transmission of CCoVs. These findings indicate that CCoV is circulating in dogs of Bangladesh. Hence, we recommend future studies on epidemiology and genetic characterization with full-genome sequencing of emerging coronaviruses in companion animals in Bangladesh.
Keywords: Bangladesh; canine; coronavirus; epidemiology; zoonotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molecular characterization of canine parvovirus and canine enteric coronavirus in diarrheic dogs on the island of St. Kitts: First report from the Caribbean region.Virus Res. 2017 Aug 15;240:154-160. doi: 10.1016/j.virusres.2017.08.008. Epub 2017 Aug 26. Virus Res. 2017. PMID: 28847699 Free PMC article.
-
Molecular Screening and Characterization of Canine Coronavirus Types I and II Strains from Domestic Dogs in Southern Italy, 2019-2021.Transbound Emerg Dis. 2024 Apr 18;2024:7272785. doi: 10.1155/2024/7272785. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303143 Free PMC article.
-
Etiology and genetic evolution of canine coronavirus circulating in five provinces of China, during 2018-2019.Microb Pathog. 2020 Aug;145:104209. doi: 10.1016/j.micpath.2020.104209. Epub 2020 Apr 18. Microb Pathog. 2020. PMID: 32311431 Free PMC article.
-
Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins.Viruses. 2014 Aug 22;6(8):3363-76. doi: 10.3390/v6083363. Viruses. 2014. PMID: 25153347 Free PMC article. Review.
-
An update on canine coronaviruses: viral evolution and pathobiology.Vet Microbiol. 2008 Dec 10;132(3-4):221-34. doi: 10.1016/j.vetmic.2008.06.007. Epub 2008 Jun 12. Vet Microbiol. 2008. PMID: 18635322 Free PMC article. Review.
Cited by
-
Comparative Examination of Feline Coronavirus and Canine Coronavirus Effects on Extracellular Vesicles Acquired from A-72 Canine Fibrosarcoma Cell Line.Vet Sci. 2025 May 15;12(5):477. doi: 10.3390/vetsci12050477. Vet Sci. 2025. PMID: 40431570 Free PMC article.
References
-
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., et al. Ninth Report of the International Committee on Taxonomy of Viruses. Springer Science & Business Media; Berlin/Heidelberg, Germany: 2011. Virus taxonomy: Classification and nomenclature of viruses; pp. 806–828.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

