Implications of the Immune Polymorphisms of the Host and the Genetic Variability of SARS-CoV-2 in the Development of COVID-19
- PMID: 35062298
- PMCID: PMC8778858
- DOI: 10.3390/v14010094
Implications of the Immune Polymorphisms of the Host and the Genetic Variability of SARS-CoV-2 in the Development of COVID-19
Abstract
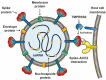
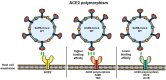
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is responsible for the current pandemic affecting almost all countries in the world. SARS-CoV-2 is the agent responsible for coronavirus disease 19 (COVID-19), which has claimed millions of lives around the world. In most patients, SARS-CoV-2 infection does not cause clinical signs. However, some infected people develop symptoms, which include loss of smell or taste, fever, dry cough, headache, severe pneumonia, as well as coagulation disorders. The aim of this work is to report genetic factors of SARS-CoV-2 and host-associated to severe COVID-19, placing special emphasis on the viral entry and molecules of the immune system involved with viral infection. Besides this, we analyze SARS-CoV-2 variants and their structural characteristics related to the binding to polymorphic angiotensin-converting enzyme type 2 (ACE2). Additionally, we also review other polymorphisms as well as some epigenetic factors involved in the immunopathogenesis of COVID-19. These factors and viral variability could explain the increment of infection rate and/or in the development of severe COVID-19.
Keywords: ACE2 overexpression; SARS-CoV-2 variant; genetic susceptibility; immune polymorphism; severe COVID-19.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Variants in ACE2; potential influences on virus infection and COVID-19 severity.Infect Genet Evol. 2021 Jun;90:104773. doi: 10.1016/j.meegid.2021.104773. Epub 2021 Feb 17. Infect Genet Evol. 2021. PMID: 33607284 Free PMC article. Review.
-
Susceptibilities of Human ACE2 Genetic Variants in Coronavirus Infection.J Virol. 2022 Jan 12;96(1):e0149221. doi: 10.1128/JVI.01492-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668773 Free PMC article.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
SARS-CoV-2 Entry Related Viral and Host Genetic Variations: Implications on COVID-19 Severity, Immune Escape, and Infectivity.Int J Mol Sci. 2021 Mar 17;22(6):3060. doi: 10.3390/ijms22063060. Int J Mol Sci. 2021. PMID: 33802729 Free PMC article. Review.
-
Host factors: Implications in immunopathogenesis of COVID-19.Pathol Res Pract. 2021 Dec;228:153647. doi: 10.1016/j.prp.2021.153647. Epub 2021 Oct 12. Pathol Res Pract. 2021. PMID: 34749207 Free PMC article. Review.
Cited by
-
Rapid SARS-CoV-2 Intra-Host and Within-Household Emergence of Novel Haplotypes.Viruses. 2022 Feb 15;14(2):399. doi: 10.3390/v14020399. Viruses. 2022. PMID: 35215992 Free PMC article.
-
Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection.Diagnostics (Basel). 2022 Nov 16;12(11):2824. doi: 10.3390/diagnostics12112824. Diagnostics (Basel). 2022. PMID: 36428884 Free PMC article.
-
SARS-CoV-2 Binding and Neutralization Properties of Peptides Derived from N-Terminus of Human ACE2.Int J Mol Sci. 2023 May 5;24(9):8269. doi: 10.3390/ijms24098269. Int J Mol Sci. 2023. PMID: 37175976 Free PMC article.
-
Innate Immune Gene Polymorphisms and COVID-19 Prognosis.Viruses. 2023 Aug 22;15(9):1784. doi: 10.3390/v15091784. Viruses. 2023. PMID: 37766191 Free PMC article.
-
HLA Variation and SARS-CoV-2 Specific Antibody Response.Viruses. 2023 Mar 31;15(4):906. doi: 10.3390/v15040906. Viruses. 2023. PMID: 37112884 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

