Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments
- PMID: 35062312
- PMCID: PMC8779076
- DOI: 10.3390/v14010108
Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments
Abstract
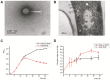
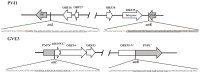
Viruses are far more abundant than cellular microorganisms in the marine ecosystem. However, very few viruses have so far been isolated from marine sediments, especially hydrothermal vent sediments, hindering the understanding of the biology and ecological functions of these tiny organisms. Here, we report the isolation and characterization of a temperate bacteriophage, named PVJ1, which infects Psychrobacillus from a hydrothermal vent field in Okinawa Trough. PVJ1 belongs to the Myoviridae family of the order Caudovirales. The tailed phage possesses a 53,187 bp linear dsDNA genome, with 84 ORFs encoding structural proteins, genome replication, host lysis, etc. in a modular pattern. The phage genome is integrated into the host chromosome near the 3'-end of deoD, a gene encoding purine nucleoside phosphorylase (PNP). The phage integration does not appear to disrupt the function of PNP. The phage DNA is packaged by the headful mechanism. Release of PVJ1 from the host cell was drastically enhanced by treatment with mitomycin C. Phages encoding an MCP sharing significant similarity (≥70% identical amino acids) with that of PVJ1 are widespread in diverse environments, including marine and freshwater sediments, soils, artificial ecosystems, and animal intestines, and primarily infect Firmicutes. These results are valuable to the understanding of the lifestyle and host interactions of bacterial viruses at the bottom of the ocean.
Keywords: Myoviridae; Psychrobacillus; bacteriophage; marine sediments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
Reply to Zrelovs et al. PVJ1 Is Not the First Tailed Temperate Phage Infecting Bacteria from Genus Psychrobacillus. Comment on "Liu et al. Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments. Viruses 2022, 14, 108".Viruses. 2022 Apr 22;14(5):866. doi: 10.3390/v14050866. Viruses. 2022. PMID: 35632608 Free PMC article.
Similar articles
-
PVJ1 Is Not the First Tailed Temperate Phage Infecting Bacteria from Genus Psychrobacillus. Comment on Liu et al. Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments. Viruses 2022, 14, 108.Viruses. 2022 Feb 28;14(3):495. doi: 10.3390/v14030495. Viruses. 2022. PMID: 35336902 Free PMC article.
-
Reply to Zrelovs et al. PVJ1 Is Not the First Tailed Temperate Phage Infecting Bacteria from Genus Psychrobacillus. Comment on "Liu et al. Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments. Viruses 2022, 14, 108".Viruses. 2022 Apr 22;14(5):866. doi: 10.3390/v14050866. Viruses. 2022. PMID: 35632608 Free PMC article.
-
Isolation and genomic characterization of Psychrobacillus isolate L3 and bacteriophage Spoks: a new phage-host pair from Antarctic soil.BMC Genomics. 2025 Apr 18;26(1):386. doi: 10.1186/s12864-025-11425-z. BMC Genomics. 2025. PMID: 40251563 Free PMC article.
-
The SPO1-related bacteriophages.Arch Virol. 2010 Oct;155(10):1547-61. doi: 10.1007/s00705-010-0783-0. Epub 2010 Aug 17. Arch Virol. 2010. PMID: 20714761 Review.
-
Tailed bacteriophages: the order caudovirales.Adv Virus Res. 1998;51:135-201. doi: 10.1016/s0065-3527(08)60785-x. Adv Virus Res. 1998. PMID: 9891587 Free PMC article. Review.
Cited by
-
PVJ1 Is Not the First Tailed Temperate Phage Infecting Bacteria from Genus Psychrobacillus. Comment on Liu et al. Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments. Viruses 2022, 14, 108.Viruses. 2022 Feb 28;14(3):495. doi: 10.3390/v14030495. Viruses. 2022. PMID: 35336902 Free PMC article.
-
Uncovering Microbial Composition of the Tissue Microenvironment in Bladder Cancer using RNA Sequencing Data.J Cancer. 2024 Mar 4;15(8):2431-2441. doi: 10.7150/jca.93055. eCollection 2024. J Cancer. 2024. PMID: 38495492 Free PMC article.
-
Polysaccharides and Their Derivatives as Potential Antiviral Molecules.Viruses. 2022 Feb 18;14(2):426. doi: 10.3390/v14020426. Viruses. 2022. PMID: 35216019 Free PMC article. Review.
-
Reply to Zrelovs et al. PVJ1 Is Not the First Tailed Temperate Phage Infecting Bacteria from Genus Psychrobacillus. Comment on "Liu et al. Isolation and Characterization of the First Temperate Virus Infecting Psychrobacillus from Marine Sediments. Viruses 2022, 14, 108".Viruses. 2022 Apr 22;14(5):866. doi: 10.3390/v14050866. Viruses. 2022. PMID: 35632608 Free PMC article.
-
Isolation and genomic characterization of Psychrobacillus isolate L3 and bacteriophage Spoks: a new phage-host pair from Antarctic soil.BMC Genomics. 2025 Apr 18;26(1):386. doi: 10.1186/s12864-025-11425-z. BMC Genomics. 2025. PMID: 40251563 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

