Discriminatory Weight of SNPs in Spike SARS-CoV-2 Variants: A Technically Rapid, Unambiguous, and Bioinformatically Validated Laboratory Approach
- PMID: 35062327
- PMCID: PMC8781109
- DOI: 10.3390/v14010123
Discriminatory Weight of SNPs in Spike SARS-CoV-2 Variants: A Technically Rapid, Unambiguous, and Bioinformatically Validated Laboratory Approach
Abstract
Background: The SARS-CoV-2 virus has assumed considerable importance during the COVID-19 pandemic. Its mutation rate is high, involving the spike (S) gene and thus there has been a rapid spread of new variants. Herein, we describe a rapid, easy, adaptable, and affordable workflow to uniquely identify all currently known variants through as few analyses. Our method only requires two conventional PCRs of the S gene and two Sanger sequencing reactions, and possibly another PCR/sequencing assay on a N gene portion to identify the B.1.160 lineage.
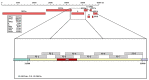
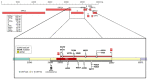
Methods: We selected an S gene 1312 bp portion containing a set of SNPs useful for discriminating all variants. Mathematical, statistical, and bioinformatic analyses demonstrated that our choice allowed us to identify all variants even without looking for all related mutations, as some of them are shared by different variants (e.g., N501Y is found in the Alpha, Beta, and Gamma variants) whereas others, that are more informative, are unique (e.g., A57 distinctive to the Alpha variant).
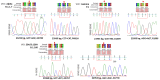
Results: A "weight" could be assigned to each mutation that may be present in the selected portion of the S gene. The method's robustness was confirmed by analyzing 80 SARS-CoV-2-positive samples.
Conclusions: Our workflow identified the variants without the need for whole-genome sequencing and with greater reliability than with commercial kits.
Keywords: COVID-19; COVID-19 variants; SARS-CoV-2; Sanger sequencing; bioinformatic validation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Stern A., Andino R. Chapter 17—Viral Evolution: It Is All About Mutations. In: Katze M., Korth M.J., Law G.L., Nathanson N., editors. Viral Pathogenesis. 3rd ed. Academic Press; Cambridge, MA, USA: 2016. pp. 233–240. - DOI
-
- [(accessed on 3 December 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

