Host-Adapted Gene Families Involved in Murine Cytomegalovirus Immune Evasion
- PMID: 35062332
- PMCID: PMC8781790
- DOI: 10.3390/v14010128
Host-Adapted Gene Families Involved in Murine Cytomegalovirus Immune Evasion
Abstract
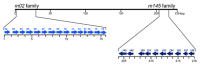
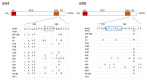
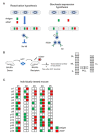
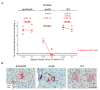
Cytomegaloviruses (CMVs) are host species-specific and have adapted to their respective mammalian hosts during co-evolution. Host-adaptation is reflected by "private genes" that have specialized in mediating virus-host interplay and have no sequence homologs in other CMV species, although biological convergence has led to analogous protein functions. They are mostly organized in gene families evolved by gene duplications and subsequent mutations. The host immune response to infection, both the innate and the adaptive immune response, is a driver of viral evolution, resulting in the acquisition of viral immune evasion proteins encoded by private gene families. As the analysis of the medically relevant human cytomegalovirus by clinical investigation in the infected human host cannot make use of designed virus and host mutagenesis, the mouse model based on murine cytomegalovirus (mCMV) has become a versatile animal model to study basic principles of in vivo virus-host interplay. Focusing on the immune evasion of the adaptive immune response by CD8+ T cells, we review here what is known about proteins of two private gene families of mCMV, the m02 and the m145 families, specifically the role of m04, m06, and m152 in viral antigen presentation during acute and latent infection.
Keywords: CD8 T cells; adoptive cell transfer; antigen presentation; antiviral protection; co-evolution; cytomegalovirus; gene family; immune evasion; latent infection/latency; memory inflation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
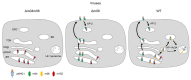
Similar articles
-
In vivo impact of cytomegalovirus evasion of CD8 T-cell immunity: facts and thoughts based on murine models.Virus Res. 2011 May;157(2):161-74. doi: 10.1016/j.virusres.2010.09.022. Epub 2010 Oct 8. Virus Res. 2011. PMID: 20933556 Review.
-
The putative natural killer decoy early gene m04 (gp34) of murine cytomegalovirus encodes an antigenic peptide recognized by protective antiviral CD8 T cells.J Virol. 2000 Feb;74(4):1871-84. doi: 10.1128/jvi.74.4.1871-1884.2000. J Virol. 2000. PMID: 10644360 Free PMC article.
-
Stochastic Episodes of Latent Cytomegalovirus Transcription Drive CD8 T-Cell "Memory Inflation" and Avoid Immune Evasion.Front Immunol. 2021 Apr 22;12:668885. doi: 10.3389/fimmu.2021.668885. eCollection 2021. Front Immunol. 2021. PMID: 33968074 Free PMC article.
-
Cytomegalovirus encodes a positive regulator of antigen presentation.J Virol. 2006 Aug;80(15):7613-24. doi: 10.1128/JVI.00723-06. J Virol. 2006. PMID: 16840340 Free PMC article.
-
Cytomegalovirus immune evasion sets the functional avidity threshold for protection by CD8 T cells.Med Microbiol Immunol. 2023 Apr;212(2):153-163. doi: 10.1007/s00430-022-00733-w. Epub 2022 Apr 1. Med Microbiol Immunol. 2023. PMID: 35364731 Free PMC article. Review.
Cited by
-
Immunotherapy of cytomegalovirus infection by low-dose adoptive transfer of antiviral CD8 T cells relies on substantial post-transfer expansion of central memory cells but not effector-memory cells.PLoS Pathog. 2023 Nov 16;19(11):e1011643. doi: 10.1371/journal.ppat.1011643. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37972198 Free PMC article.
-
Memory CD8 T Cells Protect against Cytomegalovirus Disease by Formation of Nodular Inflammatory Foci Preventing Intra-Tissue Virus Spread.Viruses. 2022 May 25;14(6):1145. doi: 10.3390/v14061145. Viruses. 2022. PMID: 35746617 Free PMC article.
-
Murine cytomegalovirus downregulates ERAAP and induces an unconventional T cell response to self.Cell Rep. 2023 Apr 25;42(4):112317. doi: 10.1016/j.celrep.2023.112317. Epub 2023 Mar 29. Cell Rep. 2023. PMID: 36995940 Free PMC article.
-
Diverse cytomegalovirus US11 antagonism and MHC-A evasion strategies reveal a tit-for-tat coevolutionary arms race in hominids.Proc Natl Acad Sci U S A. 2024 Feb 27;121(9):e2315985121. doi: 10.1073/pnas.2315985121. Epub 2024 Feb 20. Proc Natl Acad Sci U S A. 2024. PMID: 38377192 Free PMC article.
-
Cytomegalovirus inhibitors of programmed cell death restrict antigen cross-presentation in the priming of antiviral CD8 T cells.PLoS Pathog. 2024 Aug 15;20(8):e1012173. doi: 10.1371/journal.ppat.1012173. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39146364 Free PMC article.
References
-
- Davison A.J., Holton M., Dolan A., Dargan D.J., Gatherer D., Hayward G.S. Comparative genomics of primate cytomegaloviruses. In: Reddehase M.J., editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Volume 1. Caister Academic Press; Norfolk, UK: 2013. pp. 1–22.
-
- Boppana S.B., Britt W.J. Synopsis of clinical aspects of human cytomegalovirus disease. In: Reddehase M.J., editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Volume 2. Caister Academic Press; Norfolk, UK: 2013. pp. 1–25.
-
- Cannon M.J., Grosse S.D., Fowler K.B. The epidemiology and public health impact of congenital cytomegalovirus infection. In: Reddehase M.J., editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Volume 2. Caister Academic Press; Norfolk, UK: 2013. pp. 26–48.
-
- Adler S.P., Nigro G. Clinical cytomegalovirus research: Congenital infection. In: Reddehase M.J., editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Volume 2. Caister Academic Press; Norfolk, UK: 2013. pp. 55–73.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

