Actin Contributes to the Hyperexpression of Baculovirus Polyhedrin (polh) and p10 as a Component of Transcription Initiation Complex (TIC)
- PMID: 35062357
- PMCID: PMC8779803
- DOI: 10.3390/v14010153
Actin Contributes to the Hyperexpression of Baculovirus Polyhedrin (polh) and p10 as a Component of Transcription Initiation Complex (TIC)
Abstract
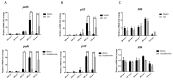
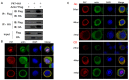
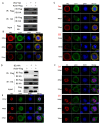
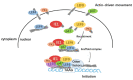
Hyperexpression of polh and p10, two very late genes, is one of the remarkable characteristics in the baculovirus life cycle. However, the mechanisms underlying the hyperexpression of these two genes are still incompletely understood. In this study, actin was identified as a highly potential binding partner of polh and p10 promoters by conducting DNA pull-down and LC-MS/MS analyses. Inhibiting actin dynamics delayed and decreased the transcription of polh and p10. Actin interacted with viral RNA polymerase and transcription regulators, and the nuclear import of viral polymerase was inhibited with the disruption of actin dynamics. Simultaneously, the high enrichment of actin in polh and p10 promoters discovered via a chromatin immunoprecipitation (ChIP) assay indicated that actin was a component of the viral polymerase TIC. Moreover, overexpression of actin surprisingly upregulated the expression of luciferase (Luc) under the control of polh and p10 promoters. Taken together, actin participated in the hyperexpression of polh and p10 as a component of TIC. These results facilitate the promotion of the expression efficiency of foreign genes in the baculovirus expression vector system (BEVS).
Keywords: BEVS; actin; baculovirus; hyperexpression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Codon Optimization-based Whole-gene Scanning Identifies Hidden Nucleotides Essential for Bombyx mori Nucleopolyhedrovirus polyhedrin Hyperexpression.J Mol Biol. 2024 Jun 15;436(12):168595. doi: 10.1016/j.jmb.2024.168595. Epub 2024 May 7. J Mol Biol. 2024. PMID: 38724003
-
Molecular mechanism responsible for the hyperexpression of baculovirus polyhedrin.Gene. 2022 Mar 10;814:146129. doi: 10.1016/j.gene.2021.146129. Epub 2021 Dec 28. Gene. 2022. PMID: 34971751 Review.
-
Specific nucleotide substitutions in the burst sequence enhance polyhedrin expression in alphabaculoviruses: improvement of baculovirus expression vectors.Appl Environ Microbiol. 2025 May 21;91(5):e0014425. doi: 10.1128/aem.00144-25. Epub 2025 Apr 7. Appl Environ Microbiol. 2025. PMID: 40192296 Free PMC article.
-
A seamless connection from the burst sequence to the start codon is essential for polyhedrin hyperexpression in alphabaculoviruses.Biochem Biophys Res Commun. 2023 Oct 30;679:1-5. doi: 10.1016/j.bbrc.2023.08.046. Epub 2023 Aug 22. Biochem Biophys Res Commun. 2023. PMID: 37651871
-
Biotechnological applications of occlusion bodies of Baculoviruses.Appl Microbiol Biotechnol. 2018 Aug;102(16):6765-6774. doi: 10.1007/s00253-018-9130-2. Epub 2018 Jun 5. Appl Microbiol Biotechnol. 2018. PMID: 29872886 Review.
References
-
- Vlak J.M., Klinkenberg A.F., Zaal K.J.M., Usmany M., Klinge-Roode E.C., Geervliet J.B.F., Roosien J., Van Lent J.W.M., Klinkenberg F. Functional studies on the p10 gene of Autographa californica nuclear polyhedrosis virus using a recombinant expressing a p10-beta-galactosidase fusion gene. Pt 4J. Gen. Virol. 1988;69:765–776. doi: 10.1099/0022-1317-69-4-765. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

