Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage
- PMID: 35062960
- PMCID: PMC8780310
- DOI: 10.1186/s12967-022-03234-9
Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage
Abstract
Background: Circulating cell-free DNA (cfDNA) can be released when myocardial damage occurs.
Methods: Here, we used the methylated CpG tandem amplification and sequencing (MCTA-seq) method for analyzing dynamic changes in heart-derived DNA in plasma samples from myocardial infarction (MI) patients.
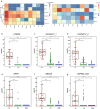
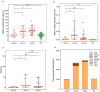
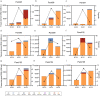
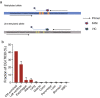
Results: We identified six CGCGCGG loci showing heart-specific hypermethylation patterns. MCTA-seq deconvolution analysis combining these loci detected heart-released cfDNA in MI patients at hospital admission, and showed that the prominently elevated total cfDNA level after percutaneous coronary intervention (PCI) was derived from both the heart and white blood cells. Furthermore, for the top marker CORO6, we developed a digital droplet PCR (ddPCR) assay that clearly detected heart damage signals in cfDNA of MI patients at hospital admission.
Conclusions: Our study provides insights into MI pathologies and developed a new ddPCR assay for detecting myocardial damage in clinical applications.
Keywords: Circulating cell-free DNA; DNA methylation; Myocardial infarction; Sequencing; ddPCR.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Comprehensive DNA methylation analysis of tissue of origin of plasma cell-free DNA by methylated CpG tandem amplification and sequencing (MCTA-Seq).Clin Epigenetics. 2019 Jun 24;11(1):93. doi: 10.1186/s13148-019-0689-y. Clin Epigenetics. 2019. PMID: 31234922 Free PMC article.
-
Digital Droplet PCR for Monitoring Tissue-Specific Cell Death Using DNA Methylation Patterns of Circulating Cell-Free DNA.Curr Protoc Mol Biol. 2019 Jun;127(1):e90. doi: 10.1002/cpmb.90. Curr Protoc Mol Biol. 2019. PMID: 31237424
-
High-throughput and affordable genome-wide methylation profiling of circulating cell-free DNA by methylated DNA sequencing (MeD-seq) of LpnPI digested fragments.Clin Epigenetics. 2021 Oct 20;13(1):196. doi: 10.1186/s13148-021-01177-4. Clin Epigenetics. 2021. PMID: 34670587 Free PMC article.
-
Computational challenges in detection of cancer using cell-free DNA methylation.Comput Struct Biotechnol J. 2021 Dec 7;20:26-39. doi: 10.1016/j.csbj.2021.12.001. eCollection 2022. Comput Struct Biotechnol J. 2021. PMID: 34976309 Free PMC article. Review.
-
Adipose cell-free DNA in diabetes.Clin Chim Acta. 2023 Jan 15;539:191-197. doi: 10.1016/j.cca.2022.12.008. Epub 2022 Dec 19. Clin Chim Acta. 2023. PMID: 36549639 Review.
Cited by
-
A Multi-Omics Approach to Defining Target Organ Injury in Youth with Primary Hypertension.bioRxiv [Preprint]. 2024 Jun 18:2024.06.17.599125. doi: 10.1101/2024.06.17.599125. bioRxiv. 2024. PMID: 38948714 Free PMC article. Preprint.
-
Genetic factors in the pathogenesis of cardio-oncology.J Transl Med. 2024 Aug 5;22(1):739. doi: 10.1186/s12967-024-05537-5. J Transl Med. 2024. PMID: 39103883 Free PMC article. Review.
-
Unlocking the secrets: the power of methylation-based cfDNA detection of tissue damage in organ systems.Clin Epigenetics. 2023 Oct 19;15(1):168. doi: 10.1186/s13148-023-01585-8. Clin Epigenetics. 2023. PMID: 37858233 Free PMC article. Review.
-
Revolution of Circulating Tumor DNA: From Bench Innovations to Bedside Implementations.Curr Issues Mol Biol. 2025 Jun 6;47(6):428. doi: 10.3390/cimb47060428. Curr Issues Mol Biol. 2025. PMID: 40699826 Free PMC article. Review.
-
RBM25 binds to and regulates alternative splicing levels of Slc38a9, Csf1, and Coro6 to affect immune and inflammatory processes in H9c2 cells.PeerJ. 2023 Nov 7;11:e16312. doi: 10.7717/peerj.16312. eCollection 2023. PeerJ. 2023. PMID: 37953772 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

