Negeviruses isolated from mosquitoes in the Brazilian Amazon
- PMID: 35062977
- PMCID: PMC8778500
- DOI: 10.1186/s12985-022-01743-z
Negeviruses isolated from mosquitoes in the Brazilian Amazon
Abstract
Background: There are several groups of viruses including Insect Specific Viruses (ISV) such as the taxon Negevirus, a group of viruses phylogenetically related to plant viruses. Negeviruses replicate in mosquito cells, but not in vertebrate cells.
Methods: Pools of hematophagous arthropods were inoculated in Vero and C6/36 cells. The cells were observed to detect possible cytopathic effect. Then, indirect immunofluorescence, RT-PCR, and nucleotide sequencing were performed.
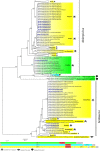
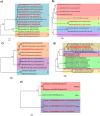
Results: Seven samples which presented negative results for flaviviruses, alphaviruses and bunyaviruses, but showed cytopathic effect in C6/36 cells were sequenced. We identified the occurrence of a variety of ISVs, most of them belonging to the taxon Negevirus: The Brejeira, Negev, Cordoba and Wallerfield viruses, including a new virus for science, tentatively named Feitosa virus.
Conclusions: We detected negeviruses in the Amazon region, including two viruses that were isolated for the first time in Brazil: Cordoba virus and the Negev virus and, a new virus for science: the Feitosa virus.
Keywords: Arboviruses; Insect-specific viruses; Negevirus.
© 2022. The Author(s).
Conflict of interest statement
The authors do hereby declare no conflict of interest.
Figures

References
-
- Auguste AJ, Carrington CVF, Forrester NL, Popov VL, Guzman H, Widen SG, Wood TG, Weaver SC. Tesh RBCharacterization of a novel Negevírus and a novel Bunyavírus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J Gen Virol. 2014;95:481–485. doi: 10.1099/vir.0.058412-0. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

