Transcriptome-scale spatial gene expression in rat arcuate nucleus during puberty
- PMID: 35063020
- PMCID: PMC8781439
- DOI: 10.1186/s13578-022-00745-2
Transcriptome-scale spatial gene expression in rat arcuate nucleus during puberty
Abstract
Background: A variety of neurons in hypothalamus undergo a complicated regulation on transcription activity of multiple genes for hypothalamic-pituitary-gonadal axis activation during pubertal development. Identification of puberty-associated cell composition and characterization of the unique transcriptional signatures across different cells are beneficial to isolation of specific neurons and advanced understanding of their functions.
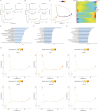
Methods: The hypothalamus of female Sprague-Dawley rats in postnatal day-25, 35 and 45 were used to define the dynamic spatial atlas of gene expression in the arcuate nucleus (ARC) by 10× Genomics Visium platform. A surface protein expressed selectively by kisspeptin neurons was used to sort neurons by flow cytometric assay in vitro. The transcriptome of the isolated cells was examined using Smart sequencing.
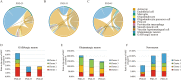
Results: Four subclusters of neurons with similar gene expression signatures in ARC were identified. Only one subcluster showed the robust expression of Kiss1, which could be isolated by a unique membrane surface biomarker Solute carrier family 18 member A3 (SLC18A3). Moreover, genes in different subclusters presenting three expression modules distinctly functioned in each pubertal stage. Different types of cells representing distinct functions on glial or neuron differentiation, hormone secretion as well as estradiol response precisely affect and coordinate with each other, resulting in a complicated regulatory network for hypothalamic-pituitary-gonadal axis initiation and modulation.
Conclusion: Our data revealed a comprehensive transcriptomic overview of ARC within different pubertal stages, which could serve as a valuable resource for the study of puberty and sexual development disorders.
Keywords: ARC; Hypothalamus; Kiss1; Puberty; Slc18a3; Spatial transcriptome.
© 2022. The Author(s).
Conflict of interest statement
All authors declare that there is no conflict of interest.
Figures




References
Grants and funding
- 81871131/National Natural Science Foundation of China
- YG2021ZD25/sponsored by the Interdisciplinary Program of Shanghai Jiao Tong University
- 2018YQN002/Hospital Level Project of Shanghai Children's Hospital
- 2017YYQ05/Cultivation Plan for Excellent Young Talents of Shanghai Children's Hospital
- 202102310443/Key Technology Research and Development Program of Shandong
LinkOut - more resources
Full Text Sources

