The combined DNA and RNA synthetic capabilities of archaeal DNA primase facilitate primer hand-off to the replicative DNA polymerase
- PMID: 35064114
- PMCID: PMC8782868
- DOI: 10.1038/s41467-022-28093-2
The combined DNA and RNA synthetic capabilities of archaeal DNA primase facilitate primer hand-off to the replicative DNA polymerase
Abstract
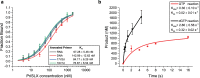
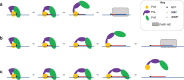
Replicative DNA polymerases cannot initiate DNA synthesis de novo and rely on dedicated RNA polymerases, primases, to generate a short primer. This primer is then extended by the DNA polymerase. In diverse archaeal species, the primase has long been known to have the ability to synthesize both RNA and DNA. However, the relevance of these dual nucleic acid synthetic modes for productive primer synthesis has remained enigmatic. In the current work, we reveal that the ability of primase to polymerize DNA serves dual roles in promoting the hand-off of the primer to the replicative DNA polymerase holoenzyme. First, it creates a 5'-RNA-DNA-3' hybrid primer which serves as an optimal substrate for elongation by the replicative DNA polymerase. Second, it promotes primer release by primase. Furthermore, modeling and experimental data indicate that primase incorporates a deoxyribonucleotide stochastically during elongation and that this switches the primase into a dedicated DNA synthetic mode polymerase.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

