A workflow for the joint modeling of longitudinal and event data in the development of therapeutics: Tools, statistical methods, and diagnostics
- PMID: 35064957
- PMCID: PMC9007602
- DOI: 10.1002/psp4.12763
A workflow for the joint modeling of longitudinal and event data in the development of therapeutics: Tools, statistical methods, and diagnostics
Abstract
Clinical trials investigate treatment endpoints that usually include measurements of pharmacodynamic and efficacy biomarkers in early-phase studies and patient-reported outcomes as well as event risks or rates in late-phase studies. In recent years, a systematic trend in clinical trial data analytics and modeling has been observed, where retrospective data are integrated into a quantitative framework to prospectively support analyses of interim data and design of ongoing and future studies of novel therapeutics. Joint modeling is an advanced statistical methodology that allows for the investigation of clinical trial outcomes by quantifying the association between baseline and/or longitudinal biomarkers and event risk. Using an exemplar data set from non-small cell lung cancer studies, we propose and test a workflow for joint modeling. It allows a modeling scientist to comprehensively explore the data, build survival models, investigate goodness-of-fit, and subsequently perform outcome predictions using interim biomarker data from an ongoing study. The workflow illustrates a full process, from data exploration to predictive simulations, for selected multivariate linear and nonlinear mixed-effects models and software tools in an integrative and exhaustive manner.
© 2022 M&S Decisions LLC. CPT: Pharmacometrics & Systems Pharmacology published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Kirill Zhudenkov, Sergey Gavrilov, Alina Sofronova, Oleg Stepanov, Nataliya Kudryashova, and Kirill Peskov are employees of M&S Decisions, LLC. Kirill Peskov is also affiliated with the Sechenov First Moscow State Medical University. Gabriel Helmlinger is an employee of Obsidian Therapeutics.
Figures

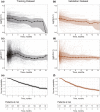


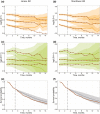
References
-
- US Department of Health and Human Services, Food and Drug Administration . Multiple endpoints in clinical trials. Guidance for industry. https://www.fda.gov/files/drugs/published/Multiple‐Endpoints‐in‐Clinical.... Published 2017. Accessed April 15, 2021.
-
- Gavrilov S, Zhudenkov K, Helmlinger G, Dunyak J, Peskov K, Aksenov S. Longitudinal tumor size and neutrophil‐to‐lymphocyte ratio are prognostic biomarkers for overall survival in patients with advanced non‐small cell lung cancer treated with durvalumab. CPT Pharmacometrics Syst Pharmacol. 2021;10(1):67‐74. 10.1002/psp4.12578 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

