Genome-wide analysis of cell-Free DNA methylation profiling with MeDIP-seq identified potential biomarkers for colorectal cancer
- PMID: 35065650
- PMCID: PMC8783473
- DOI: 10.1186/s12957-022-02487-4
Genome-wide analysis of cell-Free DNA methylation profiling with MeDIP-seq identified potential biomarkers for colorectal cancer
Abstract
Background: Colorectal cancer is the most common malignancy and the third leading cause of cancer-related death worldwide. This study aimed to identify potential diagnostic biomarkers for colorectal cancer by genome-wide plasma cell-free DNA (cfDNA) methylation analysis.
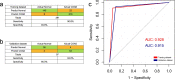
Methods: Peripheral blood from colorectal cancer patients and healthy controls was collected for cfDNA extraction. Genome-wide cfDNA methylation profiling, especially differential methylation profiling between colorectal cancer patients and healthy controls, was performed by methylated DNA immunoprecipitation coupled with high-throughput sequencing (MeDIP-seq). Logistic regression models were established, and the accuracy of this diagnostic model for colorectal cancer was verified using tissue-sourced data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) due to the lack of cfDNA methylation data in public datasets.
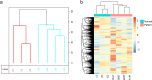
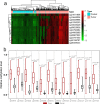
Results: Compared with the control group, 939 differentially methylated regions (DMRs) located in promoter regions were found in colorectal cancer patients; 16 of these DMRs were hypermethylated, and the remaining 923 were hypomethylated. In addition, these hypermethylated genes, mainly PRDM14, RALYL, ELMOD1, and TMEM132E, were validated and confirmed in colorectal cancer by using publicly available DNA methylation data.
Conclusions: MeDIP-seq can be used as an optimal approach for analyzing cfDNA methylomes, and 12 probes of four differentially methylated genes identified by MeDIP-seq (PRDM14, RALYL, ELMOD1, and TMEM132E) could serve as potential biomarkers for clinical application in patients with colorectal cancer.
Keywords: Biomarkers; Colorectal cancer; MeDIP-seq; cfDNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Genome-Wide Plasma Cell-Free DNA Methylation Profiling Identifies Potential Biomarkers for Lung Cancer.Dis Markers. 2019 Feb 5;2019:4108474. doi: 10.1155/2019/4108474. eCollection 2019. Dis Markers. 2019. PMID: 30867848 Free PMC article.
-
Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer.Front Genet. 2020 Dec 2;11:596078. doi: 10.3389/fgene.2020.596078. eCollection 2020. Front Genet. 2020. PMID: 33424927 Free PMC article.
-
High-throughput and affordable genome-wide methylation profiling of circulating cell-free DNA by methylated DNA sequencing (MeD-seq) of LpnPI digested fragments.Clin Epigenetics. 2021 Oct 20;13(1):196. doi: 10.1186/s13148-021-01177-4. Clin Epigenetics. 2021. PMID: 34670587 Free PMC article.
-
Analyzing the cancer methylome through targeted bisulfite sequencing.Cancer Lett. 2013 Nov 1;340(2):171-8. doi: 10.1016/j.canlet.2012.10.040. Epub 2012 Nov 28. Cancer Lett. 2013. PMID: 23200671 Free PMC article. Review.
-
Computational challenges in detection of cancer using cell-free DNA methylation.Comput Struct Biotechnol J. 2021 Dec 7;20:26-39. doi: 10.1016/j.csbj.2021.12.001. eCollection 2022. Comput Struct Biotechnol J. 2021. PMID: 34976309 Free PMC article. Review.
Cited by
-
Circulating cell-free DNA-based methylation pattern in plasma for early diagnosis of esophagus cancer.PeerJ. 2024 Jan 31;12:e16802. doi: 10.7717/peerj.16802. eCollection 2024. PeerJ. 2024. PMID: 38313016 Free PMC article. Review.
-
Gene signature developed for predicting early relapse and survival in early-stage pancreatic cancer.BJS Open. 2023 May 5;7(3):zrad031. doi: 10.1093/bjsopen/zrad031. BJS Open. 2023. PMID: 37196196 Free PMC article.
-
Coupling Immunoprecipitation with Multiplexed Digital PCR for Cell-Free DNA Methylation Detection in Small Plasma Volumes of Early-Onset Colorectal Cancer.Anal Chem. 2025 Jun 3;97(21):11259-11268. doi: 10.1021/acs.analchem.5c01361. Epub 2025 May 17. Anal Chem. 2025. PMID: 40380352 Free PMC article.
-
Age estimation using methylation-sensitive high-resolution melting (MS-HRM) in both healthy felines and those with chronic kidney disease.Sci Rep. 2021 Oct 7;11(1):19963. doi: 10.1038/s41598-021-99424-4. Sci Rep. 2021. PMID: 34620957 Free PMC article.
-
Advances in blood DNA methylation-based assay for colorectal cancer early detection: a systematic updated review.Gastroenterol Hepatol Bed Bench. 2024;17(3):225-240. doi: 10.22037/ghfbb.v17i3.2978. Gastroenterol Hepatol Bed Bench. 2024. PMID: 39308542 Free PMC article. Review.
References
-
- Insua YV, Cámara J, Vázquez EB, Fernández A, Rivera FV, Silva M, et al. Predicting outcome and therapy response in mCRC patients using an indirect method for CTCs detection by a multigene expression panel: a multicentric prospective validation study. Int J Mol Sci. 2017;18:1265. doi: 10.3390/ijms18061265. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

