Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis
- PMID: 35065702
- PMCID: PMC8841637
- DOI: 10.1016/S0140-6736(21)02724-0
Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis
Erratum in
-
Department of Error.Lancet. 2022 Oct 1;400(10358):1102. doi: 10.1016/S0140-6736(21)02653-2. Lancet. 2022. PMID: 36183727 Free PMC article. No abstract available.
Abstract
Background: Antimicrobial resistance (AMR) poses a major threat to human health around the world. Previous publications have estimated the effect of AMR on incidence, deaths, hospital length of stay, and health-care costs for specific pathogen-drug combinations in select locations. To our knowledge, this study presents the most comprehensive estimates of AMR burden to date.
Methods: We estimated deaths and disability-adjusted life-years (DALYs) attributable to and associated with bacterial AMR for 23 pathogens and 88 pathogen-drug combinations in 204 countries and territories in 2019. We obtained data from systematic literature reviews, hospital systems, surveillance systems, and other sources, covering 471 million individual records or isolates and 7585 study-location-years. We used predictive statistical modelling to produce estimates of AMR burden for all locations, including for locations with no data. Our approach can be divided into five broad components: number of deaths where infection played a role, proportion of infectious deaths attributable to a given infectious syndrome, proportion of infectious syndrome deaths attributable to a given pathogen, the percentage of a given pathogen resistant to an antibiotic of interest, and the excess risk of death or duration of an infection associated with this resistance. Using these components, we estimated disease burden based on two counterfactuals: deaths attributable to AMR (based on an alternative scenario in which all drug-resistant infections were replaced by drug-susceptible infections), and deaths associated with AMR (based on an alternative scenario in which all drug-resistant infections were replaced by no infection). We generated 95% uncertainty intervals (UIs) for final estimates as the 25th and 975th ordered values across 1000 posterior draws, and models were cross-validated for out-of-sample predictive validity. We present final estimates aggregated to the global and regional level.
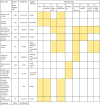
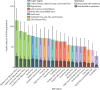
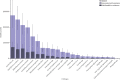
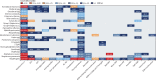
Findings: On the basis of our predictive statistical models, there were an estimated 4·95 million (3·62-6·57) deaths associated with bacterial AMR in 2019, including 1·27 million (95% UI 0·911-1·71) deaths attributable to bacterial AMR. At the regional level, we estimated the all-age death rate attributable to resistance to be highest in western sub-Saharan Africa, at 27·3 deaths per 100 000 (20·9-35·3), and lowest in Australasia, at 6·5 deaths (4·3-9·4) per 100 000. Lower respiratory infections accounted for more than 1·5 million deaths associated with resistance in 2019, making it the most burdensome infectious syndrome. The six leading pathogens for deaths associated with resistance (Escherichia coli, followed by Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa) were responsible for 929 000 (660 000-1 270 000) deaths attributable to AMR and 3·57 million (2·62-4·78) deaths associated with AMR in 2019. One pathogen-drug combination, meticillin-resistant S aureus, caused more than 100 000 deaths attributable to AMR in 2019, while six more each caused 50 000-100 000 deaths: multidrug-resistant excluding extensively drug-resistant tuberculosis, third-generation cephalosporin-resistant E coli, carbapenem-resistant A baumannii, fluoroquinolone-resistant E coli, carbapenem-resistant K pneumoniae, and third-generation cephalosporin-resistant K pneumoniae.
Interpretation: To our knowledge, this study provides the first comprehensive assessment of the global burden of AMR, as well as an evaluation of the availability of data. AMR is a leading cause of death around the world, with the highest burdens in low-resource settings. Understanding the burden of AMR and the leading pathogen-drug combinations contributing to it is crucial to making informed and location-specific policy decisions, particularly about infection prevention and control programmes, access to essential antibiotics, and research and development of new vaccines and antibiotics. There are serious data gaps in many low-income settings, emphasising the need to expand microbiology laboratory capacity and data collection systems to improve our understanding of this important human health threat.
Funding: Bill & Melinda Gates Foundation, Wellcome Trust, and Department of Health and Social Care using UK aid funding managed by the Fleming Fund.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests E Ashley reports that Lao-Oxford-Mahosot Hospital—Wellcome Trust Research Unit received financial support from the Global Research on Antimicrobial Resistance Project (GRAM) to extract and prepare data for the present manuscript. J Bielicki reports grants from the European and Developing Countries Clinical Trials Partnership, Horizon 2020, and Swiss National Science Foundation, and a contract from the National Institute for Health Research (NIHR), outside of the submitted work; and consulting fees from Shionogi and Sandoz and speaking fees from Pfizer and Sandoz, outside the submitted work. C Carvalheiro reports financial support for the present manuscript from the Global Antibiotic Research and Development Partnership, who provided payments to Fundação de Apoio ao Ensino, Pesquisa e Assistência of the Clinical Hospital of the Faculty of Medicine of Ribeirão Preto, University of São Paulo, Brazil. S Dunachie reports financial support for the present manuscript from UL Flemming Fund at the Department of Health and Social Care, the Bill & Melinda Gates Foundation, and the Wellcome Trust; a paid membership role for the Wellcome Trust Vaccines Advisory Selection Panel Vaccines and AMR in November, 2019; and an unpaid role as an expert adviser to WHO's Global Antimicrobrial Resistance Surveillance System, from November, 2018 onwards, outside the submitted work. A Haselbeck reports support for the present manuscript from the Bill & Melinda Gates Foundation (OPP1205877). C Lim was supported by the Wellcome Trust Training Fellowship between September, 2017 and March 2020 (206736/Z/17/Z), outside the submitted work. M Mussi-Pinhata reports support for the present manuscript from research from grant funding from Fondazione PENTA—Onlus and the Clinical Trial Manager Global Antibiotic R&D Partnership (GARDP). P Newton reports support for the present manuscript from research grant funding from the Wellcome Trust. J Robotham is a member of the UK Government Advisory Committee on Antimicrobial Prescribing Resistance and Healthcare Associated Infections, outside the submitted work. J Scott reports that the London School of Hygiene & Tropical Medicine (LSHTM) received financial support from Emory University to support CHAMPS projects in Ethiopia for the present manuscript; reports a paid fellowship from the Wellcome Trust, research grants from Gavi, the Vaccine Alliance, and NIHR paid to LSHTM, and an African research leader fellowship paid to LSHTM by the Medical Research Council, outside the submitted work; and reports being a member of the data safety and monitoring board for PATH Vaccines Solutions for SII PCV10 in The Gambia. J Sifuentes-Osornio reports financial support from Oxford University for the present manuscript; research grants from Oxford, CONACYT, Sanofi, and Novartis, outside of the study; consulting fees from Senosiain and speaker fees from Merck, outside of the study; and membership of the Sanofi advisory board of COVID-19 Vaccine Development, which is currently in progress, outside of the study. A J Stewardson reports grants or contracts from Merck, Sharp, & Dohme paid to Monash University, Melbourne, outside of the study. P Turner reports grants, consulting fees, and support for attending meetings or travel from Wellcome Trust, outside the study. H van Doorn reports grants or contracts from the University of Oxford and is the principal investigator for the Fleming Fund pilot grant; and he is a board member of Wellcome Trust's Surveillance and Epidemiology of Drug Resistant Infections. T Walsh reports financial support from the Bill & Melinda Gates Foundation for the BARNARDS (neonatal sepsis and mortality) study for the present manuscript. All other authors declare no competing interests.
Figures








Comment in
-
Global burden of antimicrobial resistance: essential pieces of a global puzzle.Lancet. 2022 Jun 25;399(10344):2346-2347. doi: 10.1016/S0140-6736(22)00935-7. Lancet. 2022. PMID: 35753334 No abstract available.
-
Global burden of antimicrobial resistance: essential pieces of a global puzzle.Lancet. 2022 Jun 25;399(10344):2347. doi: 10.1016/S0140-6736(22)00940-0. Lancet. 2022. PMID: 35753335 No abstract available.
-
Global burden of antimicrobial resistance: essential pieces of a global puzzle.Lancet. 2022 Jun 25;399(10344):2347-2348. doi: 10.1016/S0140-6736(22)00939-4. Lancet. 2022. PMID: 35753336 No abstract available.
-
Global burden of antimicrobial resistance: essential pieces of a global puzzle.Lancet. 2022 Jun 25;399(10344):2348. doi: 10.1016/S0140-6736(22)00944-8. Lancet. 2022. PMID: 35753337 No abstract available.
-
Global burden of antimicrobial resistance: essential pieces of a global puzzle.Lancet. 2022 Jun 25;399(10344):2348-2349. doi: 10.1016/S0140-6736(22)00943-6. Lancet. 2022. PMID: 35753338 No abstract available.
-
Nurses: an underused, vital asset against drug-resistant infections.Lancet. 2022 Sep 3;400(10354):729. doi: 10.1016/S0140-6736(22)01531-8. Lancet. 2022. PMID: 36058218 No abstract available.
References
-
- O'Neill J. Review on Antimicrobial Resistance; London: 2016. Tackling drug-resistant infections globally: final report and recommendations.
-
- O'Neill J. Review on Antimicrobial Resistance; London: 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations.
-
- National Office for Animal Health . National Office for Animal Health; Middlesex: 2016. NOAH response to final O'Neill AMR review report July 2016.
-
- WHO Antimicrobial resistance. 2021. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

