Genetic context of oncogenic drivers dictates vascular sarcoma development in aP2-Cre mice
- PMID: 35066877
- PMCID: PMC9007915
- DOI: 10.1002/path.5873
Genetic context of oncogenic drivers dictates vascular sarcoma development in aP2-Cre mice
Abstract
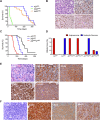
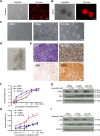
Angiosarcomas are aggressive vascular sarcomas that arise from endothelial cells and have an extremely poor prognosis. Because of the rarity of angiosarcomas, knowledge of molecular drivers and optimized treatment strategies is lacking, highlighting the need for in vivo models to study the disease. Previously, we generated genetically engineered mouse models of angiosarcoma driven by aP2-Cre-mediated biallelic loss of Dicer1 or conditional activation of KrasG12D with Cdkn2a loss that histologically and genetically resemble human tumors. In the present study, we found that DICER1 functions as a potent tumor suppressor and its deletion, in combination with either KRASG12D expression or Cdkn2a loss, is associated with angiosarcoma development. Independent of the genetic driver, the mTOR pathway was activated in all murine angiosarcoma models. Direct activation of the mTOR pathway by conditional deletion of Tsc1 with aP2-Cre resulted in tumors that resemble intermediate grade human kaposiform hemangioendotheliomas, indicating that mTOR activation was not sufficient to drive the malignant angiosarcoma phenotype. Genetic dissection of the spectrum of vascular tumors identified genes specifically regulated in the aggressive murine angiosarcomas that are also enriched in human angiosarcoma. The genetic dissection driving the transition across the malignant spectrum of endothelial sarcomas provides an opportunity to identify key determinants of the malignant phenotype, novel therapies for angiosarcoma, and novel in vivo models to further explore angiosarcoma pathogenesis. © 2022 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: DICER1; TSC1; angiosarcoma; kaposiform hemangioendothelioma; sarcoma.
© 2022 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Figures




References
-
- Dasgupta R, Fishman SJ. ISSVA classification. Semin Pediatr Surg 2014; 23: 158–161. - PubMed
-
- Putra J, Gupta A. Kaposiform haemangioendothelioma: a review with emphasis on histological differential diagnosis. Pathology 2017; 49: 356–362. - PubMed
-
- Tanas MR, Sboner A, Oliveira AM, et al. Identification of a disease‐defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med 2011; 3: 98ra82. - PubMed
-
- Florou V, Wilky BA. Current management of angiosarcoma: recent advances and lessons from the past. Curr Treat Options Oncol 2021; 22: 61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

