SOX2 mediates metabolic reprogramming of prostate cancer cells
- PMID: 35067686
- PMCID: PMC8858874
- DOI: 10.1038/s41388-021-02157-x
SOX2 mediates metabolic reprogramming of prostate cancer cells
Erratum in
-
Correction to: SOX2 mediates metabolic reprogramming of prostate cancer cells.Oncogene. 2022 Feb;41(8):1234. doi: 10.1038/s41388-022-02228-7. Oncogene. 2022. PMID: 35145235 No abstract available.
Abstract
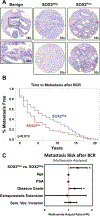
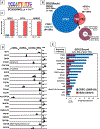
New strategies are needed to predict and overcome metastatic progression and therapy resistance in prostate cancer. One potential clinical target is the stem cell transcription factor SOX2, which has a critical role in prostate development and cancer. We thus investigated the impact of SOX2 expression on patient outcomes and its function within prostate cancer cells. Analyses of SOX2 expression among a case-control cohort of 1028 annotated tumor specimens demonstrated that SOX2 expression confers a more rapid time to metastasis and decreased patient survival after biochemical recurrence. SOX2 ChIP-Seq analyses revealed SOX2-binding sites within prostate cancer cells which differ significantly from canonical embryonic SOX2 gene targets, and prostate-specific SOX2 gene targets are associated with multiple oncogenic pathways. Interestingly, phenotypic and gene expression analyses after CRISPR-mediated deletion of SOX2 in castration-resistant prostate cancer cells, as well as ectopic SOX2 expression in androgen-sensitive prostate cancer cells, demonstrated that SOX2 promotes changes in multiple metabolic pathways and metabolites. SOX2 expression in prostate cancer cell lines confers increased glycolysis and glycolytic capacity, as well as increased basal and maximal oxidative respiration and increased spare respiratory capacity. Further, SOX2 expression was associated with increased quantities of mitochondria, and metabolomic analyses revealed SOX2-associated changes in the metabolism of purines, pyrimidines, amino acids and sugars, and the pentose phosphate pathway. Analyses of SOX2 gene targets with central functions metabolism (CERK, ECHS1, HS6SDT1, LPCAT4, PFKP, SLC16A3, SLC46A1, and TST) document significant expression correlation with SOX2 among RNA-Seq datasets derived from patient tumors and metastases. These data support a key role for SOX2 in metabolic reprogramming of prostate cancer cells and reveal new mechanisms to understand how SOX2 enables metastatic progression, lineage plasticity, and therapy resistance. Further, our data suggest clinical opportunities to exploit SOX2 as a biomarker for staging and imaging, as well as a potential pharmacologic target.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



References
-
- Wegner M All purpose Sox: The many roles of Sox proteins in gene expression. The international journal of biochemistry & cell biology 2010; 42: 381–390. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

