Skeletal muscle transcriptome response to a bout of endurance exercise in physically active and sedentary older adults
- PMID: 35068187
- PMCID: PMC8897039
- DOI: 10.1152/ajpendo.00378.2021
Skeletal muscle transcriptome response to a bout of endurance exercise in physically active and sedentary older adults
Abstract
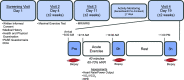
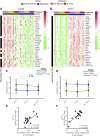
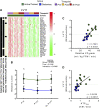
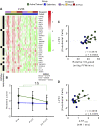
Age-related declines in cardiorespiratory fitness and physical function are mitigated by regular endurance exercise in older adults. This may be due, in part, to changes in the transcriptional program of skeletal muscle following repeated bouts of exercise. However, the impact of chronic exercise training on the transcriptional response to an acute bout of endurance exercise has not been clearly determined. Here, we characterized baseline differences in muscle transcriptome and exercise-induced response in older adults who were active/endurance trained or sedentary. RNA-sequencing was performed on vastus lateralis biopsy specimens obtained before, immediately after, and 3 h following a bout of endurance exercise (40 min of cycling at 60%-70% of heart rate reserve). Using a recently developed bioinformatics approach, we found that transcript signatures related to type I myofibers, mitochondria, and endothelial cells were higher in active/endurance-trained adults and were associated with key phenotypic features including V̇o2peak, ATPmax, and muscle fiber proportion. Immune cell signatures were elevated in the sedentary group and linked to visceral and intermuscular adipose tissue mass. Following acute exercise, we observed distinct temporal transcriptional signatures that were largely similar among groups. Enrichment analysis revealed catabolic processes were uniquely enriched in the sedentary group at the 3-h postexercise timepoint. In summary, this study revealed key transcriptional signatures that distinguished active and sedentary adults, which were associated with difference in oxidative capacity and depot-specific adiposity. The acute response signatures were consistent with beneficial effects of endurance exercise to improve muscle health in older adults irrespective of exercise history and adiposity.NEW & NOTEWORTHY Muscle transcript signatures associated with oxidative capacity and immune cells underlie important phenotypic and clinical characteristics of older adults who are endurance trained or sedentary. Despite divergent phenotypes, the temporal transcriptional signatures in response to an acute bout of endurance exercise were largely similar among groups. These data provide new insight into the transcriptional programs of aging muscle and the beneficial effects of endurance exercise to promote healthy aging in older adults.
Keywords: RNA-seq; cardiorespiratory fitness; endothelial cell; mitochondria.
Conflict of interest statement
P.M.C. is a consultant for Astellas/Mitobridge, Incorporated. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures




References
-
- Abellan van Kan G, Rolland Y, Andrieu S, Bauer J, Beauchet O, Bonnefoy M, Cesari M, Donini LM, Gillette Guyonnet S, Inzitari M, Nourhashemi F, Onder G, Ritz P, Salva A, Visser M, Vellas B. Gait speed at usual pace as a predictor of adverse outcomes in community-dwelling older people an International Academy on Nutrition and Aging (IANA) Task Force. J Nutr Health Aging 13: 881–889, 2009. doi:10.1007/s12603-009-0246-z. - DOI - PubMed
-
- Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol (1985) 112: 1625–1636, 2012. doi:10.1152/japplphysiol.00435.2011. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

