Transcriptome Profiling Based on Larvae at Different Time Points After Hatching Provides a Core Set of Gene Resource for Understanding the Metabolic Mechanisms of the Brood-Care Behavior in Octopus ocellatus
- PMID: 35069236
- PMCID: PMC8777255
- DOI: 10.3389/fphys.2021.762681
Transcriptome Profiling Based on Larvae at Different Time Points After Hatching Provides a Core Set of Gene Resource for Understanding the Metabolic Mechanisms of the Brood-Care Behavior in Octopus ocellatus
Abstract
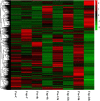
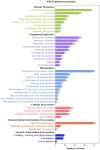
The metabolic processes of organisms are very complex. Each process is crucial and affects the growth, development, and reproduction of organisms. Metabolism-related mechanisms in Octopus ocellatus behaviors have not been widely studied. Brood-care is a common behavior in most organisms, which can improve the survival rate and constitution of larvae. Octopus ocellatus carried out this behavior, but it was rarely noticed by researchers before. In our study, 3,486 differentially expressed genes (DEGs) were identified based on transcriptome analysis of O. ocellatus. We identify metabolism-related DEGs using GO and KEGG enrichment analyses. Then, we construct protein-protein interaction networks to search the functional relationships between metabolism-related DEGs. Finally, we identified 10 hub genes related to multiple gene functions or involved in multiple signal pathways and verified them using quantitative real-time polymerase chain reaction (qRT-PCR). Protein-protein interaction networks were first used to study the effects of brood-care behavior on metabolism in the process of growing of O. ocellatus larvae, and the results provide us valuable genetic resources for understanding the metabolic processes of invertebrate larvae. The data lay a foundation for further study the brood-care behavior and metabolic mechanisms of invertebrates.
Keywords: Octopus ocellatus; brood-care behavior; metabolism; protein–protein interaction networks; transcriptome.
Copyright © 2022 Bao, Liu, Yu, Li, Cui, Wang, Feng, Xu, Sun, Li, Li and Yang.
Conflict of interest statement
BL was employed by the company Yantai Haiyu Marine Science and Technology Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures





References
-
- Arkhipkin A. I. (1985). Reproductive system structure, development and function in cephalopods with a new general scale for maturity stages. J. Northw. Atl. Fish. Sci. 12 63–74.
-
- Baeza J. A., Fernández M. (2002). Active brood care in Cancer setosus (Crustacea: Decapoda): the relationship between female behaviour, embryo oxygen consumption and the cost of brooding. Funct. Ecol. 16 241–251.
-
- Baldridge A. K., Lodge D. M. (2013). Intraguild predation between spawning smallmouth bass (Micropterus dolomieu) and nest-raiding crayfish (Orconectes rusticus): implications for bass nesting success. Freshw. Biol. 58 2355–2365.
LinkOut - more resources
Full Text Sources

