Deriving the Bidomain Model of Cardiac Electrophysiology From a Cell-Based Model; Properties and Comparisons
- PMID: 35069265
- PMCID: PMC8782150
- DOI: 10.3389/fphys.2021.811029
Deriving the Bidomain Model of Cardiac Electrophysiology From a Cell-Based Model; Properties and Comparisons
Abstract
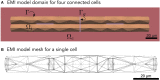
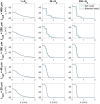
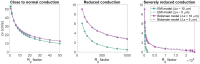
The bidomain model is considered to be the gold standard for numerical simulation of the electrophysiology of cardiac tissue. The model provides important insights into the conduction properties of the electrochemical wave traversing the cardiac muscle in every heartbeat. However, in normal resolution, the model represents the average over a large number of cardiomyocytes, and more accurate models based on representations of all individual cells have therefore been introduced in order to gain insight into the conduction properties close to the myocytes. The more accurate model considered here is referred to as the EMI model since both the extracellular space (E), the cell membrane (M) and the intracellular space (I) are explicitly represented in the model. Here, we show that the bidomain model can be derived from the cell-based EMI model and we thus reveal the close relation between the two models, and obtain an indication of the error introduced in the approximation. Also, we present numerical simulations comparing the results of the two models and thereby highlight both similarities and differences between the models. We observe that the deviations between the solutions of the models become larger for larger cell sizes. Furthermore, we observe that the bidomain model provides solutions that are very similar to the EMI model when conductive properties of the tissue are in the normal range, but large deviations are present when the resistance between cardiomyocytes is increased.
Keywords: EMI model; bidomain model; cardiac conduction; cardiac electrophysiology; cardiac tissue models; cell-based model; numerical simulation.
Copyright © 2022 Jæger and Tveito.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
From Millimeters to Micrometers; Re-introducing Myocytes in Models of Cardiac Electrophysiology.Front Physiol. 2021 Oct 27;12:763584. doi: 10.3389/fphys.2021.763584. eCollection 2021. Front Physiol. 2021. PMID: 34777021 Free PMC article.
-
Evaluating computational efforts and physiological resolution of mathematical models of cardiac tissue.Sci Rep. 2024 Jul 23;14(1):16954. doi: 10.1038/s41598-024-67431-w. Sci Rep. 2024. PMID: 39043725 Free PMC article.
-
Properties of cardiac conduction in a cell-based computational model.PLoS Comput Biol. 2019 May 31;15(5):e1007042. doi: 10.1371/journal.pcbi.1007042. eCollection 2019 May. PLoS Comput Biol. 2019. PMID: 31150383 Free PMC article.
-
Numerical solution of the bidomain equations.Philos Trans A Math Phys Eng Sci. 2009 May 28;367(1895):1931-50. doi: 10.1098/rsta.2008.0306. Philos Trans A Math Phys Eng Sci. 2009. PMID: 19380319 Review.
-
Simulating the electrical behavior of cardiac tissue using the bidomain model.Crit Rev Biomed Eng. 1993;21(1):1-77. Crit Rev Biomed Eng. 1993. PMID: 8365198 Review.
Cited by
-
Deep conditional generative model for personalization of 12-lead electrocardiograms and cardiovascular risk prediction.Front Digit Health. 2025 Apr 16;7:1558589. doi: 10.3389/fdgth.2025.1558589. eCollection 2025. Front Digit Health. 2025. PMID: 40309320 Free PMC article.
-
Efficient, cell-based simulations of cardiac electrophysiology; The Kirchhoff Network Model (KNM).NPJ Syst Biol Appl. 2023 Jun 14;9(1):25. doi: 10.1038/s41540-023-00288-3. NPJ Syst Biol Appl. 2023. PMID: 37316522 Free PMC article.
-
Generating PET scan patterns in Alzheimer's by a mathematical model.PLoS One. 2024 Apr 16;19(4):e0299637. doi: 10.1371/journal.pone.0299637. eCollection 2024. PLoS One. 2024. PMID: 38625863 Free PMC article.
-
Modeling the Electrical Activity of the Heart via Transfer Functions and Genetic Algorithms.Biomimetics (Basel). 2024 May 18;9(5):300. doi: 10.3390/biomimetics9050300. Biomimetics (Basel). 2024. PMID: 38786509 Free PMC article.
-
Arrhythmogenic influence of mutations in a myocyte-based computational model of the pulmonary vein sleeve.Sci Rep. 2022 Apr 29;12(1):7040. doi: 10.1038/s41598-022-11110-1. Sci Rep. 2022. PMID: 35487957 Free PMC article.
References
-
- Agudelo-Toro A. (2012). Numerical simulations on the biophysical foundations of the neuronal extracellular space (Ph.D. thesis). Niedersächsische Staats-und Universitätsbibliothek Göttingen.
-
- Anderson R., Andrej J., Barker A., Bramwell J., Camier J.-S., Dobrev J. C. V., et al. . (2020). MFEM: a modular finite element library. Comput. Math. Appl. 81, 42–74. 10.1016/j.camwa.2020.06.009 - DOI
LinkOut - more resources
Full Text Sources

