Long-Reads-Based Metagenomics in Clinical Diagnosis With a Special Focus on Fungal Infections
- PMID: 35069461
- PMCID: PMC8770865
- DOI: 10.3389/fmicb.2021.708550
Long-Reads-Based Metagenomics in Clinical Diagnosis With a Special Focus on Fungal Infections
Abstract
Identification of the causative infectious agent is essential in the management of infectious diseases, with the ideal diagnostic method being rapid, accurate, and informative, while remaining cost-effective. Traditional diagnostic techniques rely on culturing and cell propagation to isolate and identify the causative pathogen. These techniques are limited by the ability and the time required to grow or propagate an agent in vitro and the facts that identification based on morphological traits are non-specific, insensitive, and reliant on technical expertise. The evolution of next-generation sequencing has revolutionized genomic studies to generate more data at a cheaper cost. These are divided into short- and long-read sequencing technologies, depending on the length of reads generated during sequencing runs. Long-read sequencing also called third-generation sequencing emerged commercially through the instruments released by Pacific Biosciences and Oxford Nanopore Technologies, although relying on different sequencing chemistries, with the first one being more accurate both platforms can generate ultra-long sequence reads. Long-read sequencing is capable of entirely spanning previously established genomic identification regions or potentially small whole genomes, drastically improving the accuracy of the identification of pathogens directly from clinical samples. Long-read sequencing may also provide additional important clinical information, such as antimicrobial resistance profiles and epidemiological data from a single sequencing run. While initial applications of long-read sequencing in clinical diagnosis showed that it could be a promising diagnostic technique, it also has highlighted the need for further optimization. In this review, we show the potential long-read sequencing has in clinical diagnosis of fungal infections and discuss the pros and cons of its implementation.
Keywords: diagnosis; identification; long-read sequencing; metagenomics; mycoses; pathogenic fungi.
Copyright © 2022 Hoang, Irinyi, Hu, Schwessinger and Meyer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
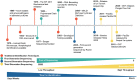

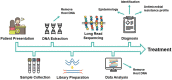


References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

