Regulatory Mechanisms of the Resistance to Common Bacterial Blight Revealed by Transcriptomic Analysis in Common Bean (Phaseolus vulgaris L.)
- PMID: 35069659
- PMCID: PMC8767069
- DOI: 10.3389/fpls.2021.800535
Regulatory Mechanisms of the Resistance to Common Bacterial Blight Revealed by Transcriptomic Analysis in Common Bean (Phaseolus vulgaris L.)
Abstract
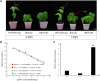
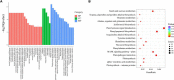
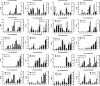
Common bean blight (CBB), primarily caused by Xanthomonas axonopodis pv. phaseoli (Xap), is one of the most destructive diseases of common bean (Phaseolus vulgaris L.). The tepary bean genotype PI 319443 displays high resistance to Xap, and the common bean genotypes HR45 and Bilu display high resistance and susceptibility to Xap, respectively. To identify candidate genes related to Xap resistance, transcriptomic analysis was performed to compare gene expression levels with Xap inoculation at 0, 24, and 48 h post inoculation (hpi) among the three genotypes. A total of 1,146,009,876 high-quality clean reads were obtained. Differentially expressed gene (DEG) analysis showed that 1,688 DEGs responded to pathogen infection in the three genotypes. Weighted gene coexpression network analysis (WGCNA) was also performed to identify three modules highly correlated with Xap resistance, in which 334 DEGs were likely involved in Xap resistance. By combining differential expression analysis and WGCNA, 139 DEGs were identified as core resistance-responsive genes, including 18 genes encoding resistance (R) proteins, 19 genes belonging to transcription factor families, 63 genes encoding proteins with oxidoreductase activity, and 33 plant hormone signal transduction-related genes, which play important roles in the resistance to pathogen infection. The expression patterns of 20 DEGs were determined by quantitative real-time PCR (qRT-PCR) and confirmed the reliability of the RNA-seq results.
Keywords: RNA-seq; WGCNA; common bacterial blight; common bean; plant resistance.
Copyright © 2022 Yang, Chang, Wang, Wang and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bakade R., Ingole K. D., Deshpande S., Pal G., Patil S. S., Bhattacharjee S., et al. (2021). Comparative transcriptome analysis of rice resistant and susceptible genotypes to Xanthomonas oryzae pv. oryzae identifies novel genes to control bacterial leaf blight. Mol. Biotechnol. 63 719–731. 10.1007/s12033-021-00338-3 - DOI - PubMed
-
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., et al. (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9 200–211. 10.1016/j.chom.2011.02.009 - DOI - PMC - PubMed
-
- Brouwer B., Winkler L., Atterberry K., Jones S., Miles C. (2016). Exploring the role of local heirloom germplasm in expanding western Washington dry bean production. Agroecol. Sust. Food 40 319–332. 10.1080/21683565.2015.1138013 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

